Origin and Evolution of Enzymes with MIO Prosthetic Group: Microbial Coevolution After the Mass Extinction Event
- PMID: 35422843
- PMCID: PMC9002059
- DOI: 10.3389/fgene.2022.851738
Origin and Evolution of Enzymes with MIO Prosthetic Group: Microbial Coevolution After the Mass Extinction Event
Abstract
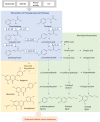
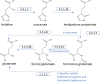
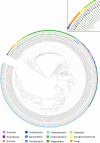
After major mass extinction events, ancient plants and terrestrial vertebrates were faced with various challenges, especially ultraviolet (UV) light. These stresses probably resulted in changes in the biosynthetic pathways, which employed the MIO (3,5-dihydro-5-methylidene-4H-imidazole-4-one)-dependent enzymes (ammonia-lyase and aminomutase), leading to enhanced accumulation of metabolites for defense against UV radiation, pathogens, and microorganisms. Up to now, the origin and evolution of genes from this superfamily have not been extensively studied. In this report, we perform an analysis of the phylogenetic relations between the members of the aromatic amino acid MIO-dependent enzymes (AAM), which demonstrate that they most probably have a common evolutionary origin from ancient bacteria. In early soil environments, numerous bacterial species with tyrosine ammonia-lyase genes (TAL; EC 4.3.1.23) developed tyrosine aminomutase (TAM; EC 5.4.3.6) activity as a side reaction for competing with their neighbors in the community. These genes also evolved into other TAL-like enzymes, such as histidine ammonia-lyase (HAL, EC 4.3.1.3) and phenylalanine ammonia-lyase (PAL; EC 4.3.1.24), in different bacterial species for metabolite production and accumulation for adaptation to adverse terrestrial environmental conditions. On the other hand, the existence of phenylalanine aminomutase (PAM; EC 5.4.3.10) and phenylalanine/tyrosine ammonia-lyase (PTAL; EC 4.3.1.25) strongly indicates the horizontal gene transfer (HGT) between bacteria, fungi, and plants in symbiotic association after acquiring the PAL gene from their ancestor.
Keywords: MIO-dependent enzymes; a mass extinction event; microbial coevolution; minimal ancestor deviation; rooted phylogenetic tree.
Copyright © 2022 Peng, Engel, Aliyu and Rudat.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Averof M., Crc W., Akam M. (1995). Insect-crustacean Relationships: Insights from Comparative Developmental and Molecular Studies. Phil. Trans. R. Soc. Lond. B 347, 293–303. 10.1098/rstb.1995.0028 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

