Whole-Genome Sequencing of Endangered Dengchuan Cattle Reveals Its Genomic Diversity and Selection Signatures
- PMID: 35422847
- PMCID: PMC9001881
- DOI: 10.3389/fgene.2022.833475
Whole-Genome Sequencing of Endangered Dengchuan Cattle Reveals Its Genomic Diversity and Selection Signatures
Abstract
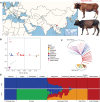
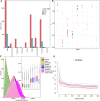
Dengchuan cattle are the only dairy yellow cattle and endangered cattle among Yunnan native cattle breeds. However, its genetic background remains unclear. Here, we performed whole-genome sequencing of ten Dengchuan cattle. Integrating our data with the publicly available data, Dengchuan cattle were observed to be highly interbred than other cattle in the dataset. Furthermore, the positive selective signals were mainly manifested in candidate genes and pathways related to milk production, disease resistance, growth and development, and heat tolerance. Notably, five genes (KRT39, PGR, KRT40, ESR2, and PRKACB) were significantly enriched in the estrogen signaling pathway. Moreover, the missense mutation in the PGR gene (c.190T > C, p.Ser64Pro) showed a homozygous mutation pattern with higher frequency (83.3%) in Dengchuan cattle. In addition, a large number of strong candidate regions matched genes and QTLs related to milk yield and composition. Our research provides a theoretical basis for analyzing the genetic mechanism underlying Dengchuan cattle with excellent lactation and adaptability, crude feed tolerance, good immune performance, and small body size and also laid a foundation for genetic breeding research of Dengchuan cattle in the future.
Keywords: Chinese cattle; genetic diversity; lactation function; population structure; selection signatures; whole-genome resequencing.
Copyright © 2022 Jin, Qu, Hanif, Zhang, Liu, Chen, Suolang, Lei and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Alain K., Karrow N. A., Thibault C., St-Pierre J., Lessard M., Bissonnette N. (2009). Osteopontin: an Early Innate Immune Marker of Escherichia coli Mastitis Harbors Genetic Polymorphisms with Possible Links with Resistance to Mastitis. BMC Genomics 10, 444. 10.1186/1471-2164-10-444 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

