A familial 3q28q29 duplication induced mild intellectual disability: case presentation and literature review
- PMID: 35422908
- PMCID: PMC8991147
A familial 3q28q29 duplication induced mild intellectual disability: case presentation and literature review
Abstract
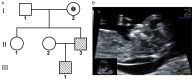
The 3q29 duplication syndrome is an uncommon imbalanced chromosomal disorder with highly variable manifestations, mainly characterized by a mild mental anomaly, eye abnormalities, and developmental delay. Only a few such cases have been reported with significant phenotypic heterogeneity. Here, we reported a case with familial 3q28q29 duplication that was 8.5 Mb in length, covering all fragments from previous reports. A series of genetic detection techniques, including karyotyping, chromosomal microarray, and fluorescence in situ hybridization, demonstrated that the rearrangement, in this case, was due to a three-chromosome translocation of the paternal grandmother of the fetus. Interestingly, only mild intellectual disability in the father and slightly thick nuchal translucency (NT) in the fetus were observed. The fetus was delivered at term and showed normal developmental milestones. Our study increased the understanding of this syndrome and highlighted the necessity and importance of the rational use of multiple genetic techniques in prenatal diagnosis.
Keywords: 3q29 duplication; SNP array; fluorescence in situ hybridization; intellectual disability.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures



References
-
- Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, Meng L, Ye Y, Wang J, Zhang M, Li Y, Wang H, Yu S, Chen F, Xie J, Jiang H, Wang W, Choy KW, Xu Z. Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med. 2016;18:940–948. - PubMed
-
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. - PMC - PubMed
-
- Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, Raca G, Ritter DI, South ST, Thorland EC, Pineda-Alvarez D, Aradhya S, Martin CL. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet Med. 2020;22:245–257. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
