SOX18-associated gene signature predicts sepsis outcome
- PMID: 35422958
- PMCID: PMC8991122
SOX18-associated gene signature predicts sepsis outcome
Abstract
Objectives: Sepsis is a critical medical condition associated with an high mortality. Currently, there are no reliable diagnostic or prognostic biomarkers to evaluate sepsis outcomes. SRY (sex-determining region on the Y chromosome)-box transcription factor 18 (SOX18) is an endothelial barrier protective protein, and a decreased level of SOX18 expression is involved in disruption of human endothelial cell barrier integrity. Over-expression of SOX18 attenuates the bacterial lipopolysaccharide (LPS)-mediated disruption of the vascular barrier and is associated with favorable prognosis. The utility of SOX18-related genes as biomarkers in sepsis is uncertain.
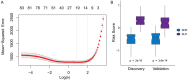
Methods: Transcriptomic analysis was used to profile the PBMC samples of patients with sepsis across two Gene Expression Omnibus (GEO) datasets with survival data. An 84-gene signature was derived from discovery datasets that correlated with SOX18 gene expression and sepsis survival.
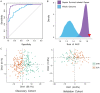
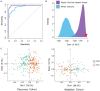
Results: Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed Th1 and Th2 cell differentiation, Cytokine-cytokine receptor interaction, and T cell receptor signaling pathways as the most significantly enriched KEGG pathways among 84 genes. A severity score based on the gene expression of 84 genes was allocated to each patient. A notable increase was detected in sepsis patients compared to healthy controls in both discovery and validation cohorts. SOX18-associated gene signature discriminated severe cases from mild cases and performed significantly better than both random 84-gene sets from whole genomes or sepsis survival-related genes. Furthermore, we obtained an 18-gene signature from screening these 84 genes in a LASSO model, which performed better in both discovery and validation cohorts.
Conclusions: Data support SOX18-associated gene signatures as a prognostic biomarker for sepsis.
Keywords: SOX18; gene signature; sepsis survival.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures


References
-
- Cohen J, Vincent JL, Adhikari NKJ, Machado FR, Angus DC, Calandra T, Jaton K, Giulieri S, Delaloye J, Opal S, Tracey K, van der Poll T, Pelfrene E. Sepsis: a roadmap for future research. Lancet Infect Dis. 2015;15:581–614. - PubMed
-
- Annane D. Sepsis clinical knowledge: a role of steroid treatment. Minerva Anestesiol. 2003;69:254–257. - PubMed
-
- Deigner HP, Kohl M. The molecular sepsis signature. Crit Care Med. 2009;37:1137–1138. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
