Coordination complexes constructed from pyrazole-acetamide and pyrazole-quinoxaline: effect of hydrogen bonding on the self-assembly process and antibacterial activity
- PMID: 35425576
- PMCID: PMC8981392
- DOI: 10.1039/d1ra09027e
Coordination complexes constructed from pyrazole-acetamide and pyrazole-quinoxaline: effect of hydrogen bonding on the self-assembly process and antibacterial activity
Abstract
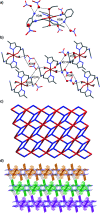
Two mononuclear coordination complexes of N-(2-aminophenyl)-2-(5-methyl-1H-pyrazol-3-yl)acetamide (L1), namely [Cd(L1)2Cl2] (C1) and [Cu(L1)2(C2H5OH)2](NO3)2 (C2) and one mononuclear complex [Fe(L2)2(H2O)2](NO3)2·2H2O (C3), obtained after in situ oxidation of L1, have been synthesized and characterized spectroscopically. As revealed by single-crystal X-ray diffraction, each coordination sphere made of two heterocycles is completed either by two chloride anions (in C1), two ethanol molecules (in C2) or two water molecules (in C3). The crystal packing analysis of C1, C2 and C3, revealed 1D and 2D supramolecular architectures, respectively, via various hydrogen bonding interactions, which are discussed in detail. Furthermore, evaluation in vitro of the ligands and their metal complexes for their antibacterial activity against Escherichia coli (ATCC 4157), Pseudomonas aeruginosa (ATCC 27853), Staphylococcus aureus (ATCC 25923) and Streptococcus fasciens (ATCC 29212) strains of bacteria, revealed outstanding results compared to chloramphenicol, a well-known antibiotic, with a normalized minimum inhibitory concentration as low as 5 μg mL-1.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
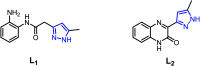
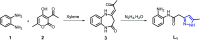







References
-
- European Centre for Disease Prevention and Control, Antibiotic Resistance: An Increasing Threat to Human Health, 2018,https://antibiotic.ecdc.europa.eu/en/publications-data/antibiotic-resist..., accessed on 18 Jan. 2022
-
- Atlanta, GA: US Centers for Disease Control and Prevention, Antibiotic Resistance Threats in the United States, 2019,https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threatsrep..., accessed on 18 Jan. 2022
-
- Geneva: World Health Organization, Antimicrobial resistance,https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance,accessed on 18 Jan. 2022
-
- Bhaumik P. K. Ghosh K. Chattopadhyay S. Synthetic strategies, crystal structures and biological activities of metal complexes with the members of azole family: A review. Polyhedron. 2021;200:115093. doi: 10.1016/j.poly.2021.115093. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

