Multi-omics Profiling Shows BAP1 Loss Is Associated with Upregulated Cell Adhesion Molecules in Uveal Melanoma
- PMID: 35426938
- PMCID: PMC9357204
- DOI: 10.1158/1541-7786.MCR-21-0657
Multi-omics Profiling Shows BAP1 Loss Is Associated with Upregulated Cell Adhesion Molecules in Uveal Melanoma
Abstract
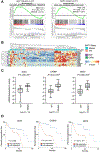
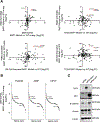
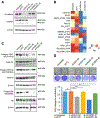
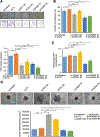
BRCA1-associated protein 1 (BAP1) is a tumor suppressor gene that is mutated in cancer, including uveal melanoma. Loss-of-function BAP1 mutations are associated with uveal melanoma metastasis and poor prognosis, but the mechanisms underlying these effects remain unclear. Upregulation of cell-cell adhesion proteins is involved with collective migration and metastatic seeding of cancer cells. Here, we show that BAP1 loss in uveal melanoma patient samples is associated with upregulated gene expression of multiple cell adhesion molecules (CAM), including E-cadherin (CDH1), cell adhesion molecule 1 (CADM1), and syndecan-2 (SDC2). Similar findings were observed in uveal melanoma cell lines and single-cell RNA-sequencing data from uveal melanoma patient samples. BAP1 reexpression in uveal melanoma cells reduced E-cadherin and CADM1 levels. Functionally, knockdown of E-cadherin decreased spheroid cluster formation and knockdown of CADM1 decreased growth of BAP1-mutant uveal melanoma cells. Together, our findings demonstrate that BAP1 regulates the expression of CAMs which may regulate metastatic traits.
Implications: BAP1 mutations and increased metastasis may be due to upregulation of CAMs.
©2022 American Association for Cancer Research.
Figures

References
-
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998;16: 1097–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

