Classification of human chronic inflammatory skin disease based on single-cell immune profiling
- PMID: 35427179
- PMCID: PMC9301819
- DOI: 10.1126/sciimmunol.abl9165
Classification of human chronic inflammatory skin disease based on single-cell immune profiling
Abstract
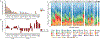
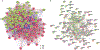
Inflammatory conditions represent the largest class of chronic skin disease, but the molecular dysregulation underlying many individual cases remains unclear. Single-cell RNA sequencing (scRNA-seq) has increased precision in dissecting the complex mixture of immune and stromal cell perturbations in inflammatory skin disease states. We single-cell-profiled CD45+ immune cell transcriptomes from skin samples of 31 patients (7 atopic dermatitis, 8 psoriasis vulgaris, 2 lichen planus (LP), 1 bullous pemphigoid (BP), 6 clinical/histopathologically indeterminate rashes, and 7 healthy controls). Our data revealed active proliferative expansion of the Treg and Trm components and universal T cell exhaustion in human rashes, with a relative attenuation of antigen-presenting cells. Skin-resident memory T cells showed the greatest transcriptional dysregulation in both atopic dermatitis and psoriasis, whereas atopic dermatitis also demonstrated recurrent abnormalities in ILC and CD8+ cytotoxic lymphocytes. Transcript signatures differentiating these rash types included genes previously implicated in T helper cell (TH2)/TH17 diatheses, segregated in unbiased functional networks, and accurately identified disease class in untrained validation data sets. These gene signatures were able to classify clinicopathologically ambiguous rashes with diagnoses consistent with therapeutic response. Thus, we have defined major classes of human inflammatory skin disease at the molecular level and described a quantitative method to classify indeterminate instances of pathologic inflammation. To make this approach accessible to the scientific community, we created a proof-of-principle web interface (RashX), where scientists and clinicians can visualize their patient-level rash scRNA-seq-derived data in the context of our TH2/TH17 transcriptional framework.
Figures




References
-
- Rachakonda TD, Schupp CW, Armstrong AW, Psoriasis prevalence among adults in the United States. J Am Acad Dermatol 70, 512–516 (2014). - PubMed
-
- Chiesa Fuxench ZC, Block JK, Boguniewicz M, Boyle J, Fonacier L, Gelfand JM, Grayson MH, Margolis DJ, Mitchell L, Silverberg JI, Schwartz L, Simpson EL, Ong PY, Atopic Dermatitis in America Study: A Cross-Sectional Study Examining the Prevalence and Disease Burden of Atopic Dermatitis in the US Adult Population. J Invest Dermatol 139, 583–590 (2019). - PubMed
-
- Simpson EL, Bieber T, Guttman-Yassky E, Beck LA, Blauvelt A, Cork MJ, Silverberg JI, Deleuran M, Kataoka Y, Lacour J-P, Kingo K, Worm M, Poulin Y, Wollenberg A, Soo Y, Graham NMH, Pirozzi G, Akinlade B, Staudinger H, Mastey V, Eckert L, Gadkari A, Stahl N, Yancopoulos GD, Ardeleanu M, Two Phase 3 Trials of Dupilumab versus Placebo in Atopic Dermatitis. N Engl J Med 375, 2335–2348 (2016). - PubMed
-
- Ferreira S, Guttman-Yassky E, Torres T, Selective JAK1 Inhibitors for the Treatment of Atopic Dermatitis: Focus on Upadacitinib and Abrocitinib. Am J Clin Dermatol 21, 783–798 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

