Cryo-EM structure of the prothrombin-prothrombinase complex
- PMID: 35427420
- PMCID: PMC9203702
- DOI: 10.1182/blood.2022015807
Cryo-EM structure of the prothrombin-prothrombinase complex
Abstract
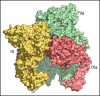
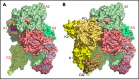
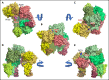
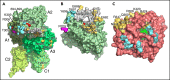
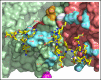
The intrinsic and extrinsic pathways of the coagulation cascade converge to a common step where the prothrombinase complex, comprising the enzyme factor Xa (fXa), the cofactor fVa, Ca2+ and phospholipids, activates the zymogen prothrombin to the protease thrombin. The reaction entails cleavage at 2 sites, R271 and R320, generating the intermediates prethrombin 2 and meizothrombin, respectively. The molecular basis of these interactions that are central to hemostasis remains elusive. We solved 2 cryogenic electron microscopy (cryo-EM) structures of the fVa-fXa complex, 1 free on nanodiscs at 5.3-Å resolution and the other bound to prothrombin at near atomic 4.1-Å resolution. In the prothrombin-fVa-fXa complex, the Gla domains of fXa and prothrombin align on a plane with the C1 and C2 domains of fVa for interaction with membranes. Prothrombin and fXa emerge from this plane in curved conformations that bring their protease domains in contact with each other against the A2 domain of fVa. The 672ESTVMATRKMHDRLEPEDEE691 segment of the A2 domain closes on the protease domain of fXa like a lid to fix orientation of the active site. The 696YDYQNRL702 segment binds to prothrombin and establishes the pathway of activation by sequestering R271 against D697 and directing R320 toward the active site of fXa. The cryo-EM structure provides a molecular view of prothrombin activation along the meizothrombin pathway and suggests a mechanism for cleavage at the alternative R271 site. The findings advance our basic knowledge of a key step of coagulation and bear broad relevance to other interactions in the blood.
© 2022 by The American Society of Hematology.
Figures


Comment in
-
Shining a light on thrombin activation.Blood. 2022 Jun 16;139(24):3451-3453. doi: 10.1182/blood.2022016537. Blood. 2022. PMID: 35708725 No abstract available.
-
What the neighbors say.J Thromb Haemost. 2022 Aug;20(8):1755. doi: 10.1111/jth.15796. J Thromb Haemost. 2022. PMID: 35859280 No abstract available.
References
-
- Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: Initiation, maintenance, and regulation. Biochemistry. 1991;30(43):10363-10370. - PubMed
-
- Mann KG. Thrombin formation. Chest. 2003;124(3 suppl):4S-10S. - PubMed
-
- Gailani D, Broze GJ Jr. Factor XI activation in a revised model of blood coagulation. Science. 1991;253(5022):909-912. - PubMed
-
- Rosing J, Tans G, Govers-Riemslag JW, Zwaal RF, Hemker HC. The role of phospholipids and factor Va in the prothrombinase complex. J Biol Chem. 1980;255(1):274-283. - PubMed
-
- Mann KG, Kalafatis M. Factor V: A combination of Dr Jekyll and Mr Hyde. Blood. 2003;101(1):20-30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

