De novo transcriptome assembly and analysis of gene expression in different tissues of moth bean (Vigna aconitifolia) (Jacq.) Marechal
- PMID: 35428206
- PMCID: PMC9013028
- DOI: 10.1186/s12870-022-03583-z
De novo transcriptome assembly and analysis of gene expression in different tissues of moth bean (Vigna aconitifolia) (Jacq.) Marechal
Abstract
Background: The underutilized species Vigna aconitifolia (Moth Bean) is an important legume crop cultivated in semi-arid conditions and is valued for its seeds for their high protein content. It is also a popular green manure cover crop that offers many agronomic benefits including nitrogen fixation and soil nutrients. Despite its economic potential, genomic resources for this crop are scarce and there is limited knowledge on the developmental process of this plant at a molecular level. In the present communication, we have studied the molecular mechanisms that regulate plant development in V. aconitifolia, with a special focus on flower and seed development. We believe that this study will greatly enrich the genomic resources for this plant in form of differentially expressed genes, transcription factors, and genic molecular markers.
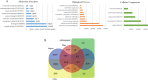
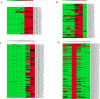
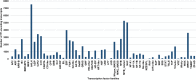
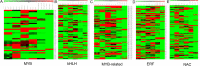
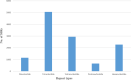
Results: We have performed the de novo transcriptome assembly using six types of tissues from various developmental stages of Vigna aconitifolia (var. RMO-435), namely, leaves, roots, flowers, pods, and seed tissue in the early and late stages of development, using the Illumina NextSeq platform. We assembled the transcriptome to get 150938 unigenes with an average length of 937.78 bp. About 79.9% of these unigenes were annotated in public databases and 12839 of those unigenes showed a significant match in the KEGG database. Most of the unigenes displayed significant differential expression in the late stages of seed development as compared with leaves. We annotated 74082 unigenes as transcription factors and identified 12096 simple sequence repeats (SSRs) in the genic regions of V.aconitifolia. Digital expression analysis revealed specific gene activities in different tissues which were validated using Real-time PCR analysis.
Conclusions: The Vigna aconitifolia transcriptomic resources generated in this study provide foundational resources for gene discovery with respect to various developmental stages. This study provides the first comprehensive analysis revealing the genes involved in molecular as well as metabolic pathways that regulate seed development and may be responsible for the unique nutritive values of moth bean seeds. Hence, this study would serve as a foundation for characterization of candidate genes which would not only provide novel insights into understanding seed development but also provide resources for improved moth bean and related species genetic enhancement.
Keywords: Developmental stages; Differential expression; Transcriptome assembly; Vigna aconitifolia.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Katoch R. Ricebean. 2020.
-
- Bhadkaria A, Srivastava N, Bhagyawant SS. A prospective of underutilized legume moth bean (Vigna aconitifolia (Jacq.) Marechàl): Phytochemical profiling, bioactive compounds and in vitro pharmacological studies. Food Biosci. 2021;42:101088. doi: 10.1016/j.fbio.2021.101088. - DOI
-
- Shah TI, Rai AP, M.a A. Relationship of Phosphorus Fractions with Soil Properties in Mothbean Growing Acid Soils of North Western Indian Himalayas. Communications in Soil Science And Plant Analysis. 2019;50(9):1192-8. 10.1080/00103624.2019.1604730.
-
- Verma N, Sehrawat KD, Ahlawat A, Sehrawat AR. International Journal of Cell Science and Biotechnology Legumes: The Natural Products for Industrial and Medicinal Importance-A Review. 6. Int J Cell Sci Biotechnol. 2017;6:5.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

