African derived phytocompounds may interfere with SARS-CoV-2 RNA capping machinery via inhibition of 2'-O-ribose methyltransferase: An in silico perspective
- PMID: 35431328
- PMCID: PMC9002684
- DOI: 10.1016/j.molstruc.2022.133019
African derived phytocompounds may interfere with SARS-CoV-2 RNA capping machinery via inhibition of 2'-O-ribose methyltransferase: An in silico perspective
Abstract
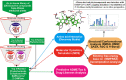
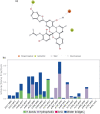
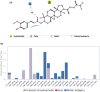
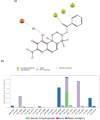
Despite the ongoing vaccination against the life-threatening COVID-19, there is need for viable therapeutic interventions. The S-adenosyl-l-Methionine (SAM) dependent 2-O'-ribose methyltransferase (2'-O-MTase) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) presents a therapeutic target against COVID-19 infection. In a bid to profile bioactive principles from natural sources, a custom-made library of 226 phytochemicals from African medicinal plants with especially anti-malarial activity was screened for direct interactions with SARS-CoV-2 2'-O-MTase (S2RMT) using molecular docking and molecular dynamics (MD) simulations as well as binding free energies methods. Based on minimal binding energy lower than sinefungin (a reference methyl-transferase inhibitor) and binding mode analysis at the catalytic site of S2RMT, a list of 26 hit phytocompounds was defined. The interaction of these phytocompounds was compared with the 2'-O-MTase of SARS-CoV and MERS-CoV. Among these compounds, the lead phytocompounds (LPs) viz: mulberrofuran F, 24-methylene cycloartenol, ferulate, 3-benzoylhosloppone and 10-hydroxyusambarensine interacted strongly with the conserved KDKE tetrad within the substrate binding pocket of the 2'-O-MTase of the coronavirus strains which is critical for substrate binding. The thermodynamic parameters analyzed from the MD simulation trajectories of the LPs-S2RMT complexes presented an eminent structural stability and compactness. These LPs demonstrated favorable druggability and in silico ADMET properties over a diverse array of molecular computing descriptors. The LPs show promising prospects in the disruption of S2RMT capping machinery in silico. However, these LPs should be validated via in vitro and in vivo experimental models.
Keywords: 2-O’-ribosemethyltransferase; Coronavirus; Molecular docking; Molecular dynamics; Mulberrofuran F; Phytochemicals; SARS-CoV-2.
© 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









References
-
- World Health Organization . World Health Organization; 2021. Coronavirus Disease (COVID-19) Weekly Epidemiological. https://www.who.int/publications/m/item/weekly-epidemiological-update-on....
-
- Zhou P., YANG X., wANG X., HU B., w Z.H.A.N.G., Hr Si, y zHu, li B., cl HuanG, cHen H.D., cHen J., luo y, Guo H., rD JianG, liu M.Q., y cHen, Xr SHen, WanG X., zHenG X.S., zHao K., cHen Q.J., DenG F., ll liu, yan B., zHan F.X., yy WanG, Xiao G.F., zl SHi. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

