COVID-19 vaccines: their effectiveness against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its emerging variants
- PMID: 35431535
- PMCID: PMC8991668
- DOI: 10.1186/s42269-022-00787-z
COVID-19 vaccines: their effectiveness against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its emerging variants
Abstract
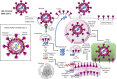
Background: The world has been suffering from the COVID-19 pandemic (officially declared by WHO in March 2020), caused by the severe acute respiratory β-coronavirus 2 (SARS-CoV-2) since the last week of December 2019. The disease was initially designated as a Public Health Emergency of International Concern on January 30, 2020. In order to protect the health of mass public, an array of research on drugs and vaccines against SARS-CoV-2 has been conducted globally. However, the emerging variants of SARS-CoV-2, i.e., Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) variants which evolved in late 2020 and the Omicron variant (B.1.1.529) which emerged in November 2021 along with its subvariant BA.2 which was first identified in India and South Africa in late December 2021, have raised the doubt about the efficiency of the currently used vaccines especially in terms of the consistent potential to produce neutralizing antibodies targeting the viral spike (S) protein.
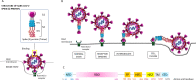
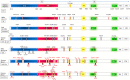
Main body of the abstract: The present review discussed the functional details of major vaccines regarding their efficiency against such variants during the pandemic. Overall, the mRNA vaccines have shown around 94% effectiveness; the adenovector vaccine showed approximately 70% efficacy, whereas Sputnik V vaccines showed around 92% effectiveness; the inactivated whole-virus vaccine CoronaVac/PiCoVacc and BBIBP-CorV showed a varying effectiveness of 65-86% according to the geographic locations; the subunit vaccine NVX-CoV2373 has shown 60-89% effectiveness along with the global regions against the wild-type SARS-CoV-2 strain. However, reduced effectiveness of these vaccines against the SARS-CoV-2 variants was noticed which is suggestive for the further administration of booster dose.
Short conclusion: Maximum variants of SARS-CoV-2 emerged during the second wave of COVID-19; and extensive studies on the viral genomic sequences from all geographical locations around the world have been conducted by an array of groups to assess the possible occurrence of mutations(s) specially within the receptor binding domain of the viral spike (S) protein. Mutational similarities and the new or critical mutations within all variants have been clearly identified so far. The study of effectiveness of the currently used vaccines is also ongoing. The persistence of memory B cell action and the other immune components as well as the administration of booster dose is expected to mitigate the disease.
Keywords: COVID-19; Mutations; Neutralizing antibodies; SARS-CoV-2; Vaccine effectiveness; Vaccines; Variants.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsThe authors declare that they have no conflict of interest.
Figures
Similar articles
-
The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem.J Biol Regul Homeost Agents. 2021 Jan-Feb;35(1):1-4. doi: 10.23812/21-3-E. J Biol Regul Homeost Agents. 2021. PMID: 33377359
-
COVID-19 Pandemic and Vaccines Update on Challenges and Resolutions.Front Cell Infect Microbiol. 2021 Sep 10;11:690621. doi: 10.3389/fcimb.2021.690621. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34568087 Free PMC article. Review.
-
Immune response of booster doses of BBIBP-CORV vaccines against the variants of concern of SARS-CoV-2.J Clin Virol. 2022 Jun;150-151:105161. doi: 10.1016/j.jcv.2022.105161. Epub 2022 Apr 12. J Clin Virol. 2022. PMID: 35439702 Free PMC article.
-
Covid-19 vaccines and variants of concern: A review.Rev Med Virol. 2022 Jul;32(4):e2313. doi: 10.1002/rmv.2313. Epub 2021 Nov 9. Rev Med Virol. 2022. PMID: 34755408 Free PMC article. Review.
-
Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness.Vaccines (Basel). 2022 Oct 19;10(10):1751. doi: 10.3390/vaccines10101751. Vaccines (Basel). 2022. PMID: 36298616 Free PMC article. Review.
Cited by
-
COVID-19 Vaccines: An Updated Overview of Different Platforms.Bioengineering (Basel). 2022 Nov 19;9(11):714. doi: 10.3390/bioengineering9110714. Bioengineering (Basel). 2022. PMID: 36421115 Free PMC article. Review.
-
What Should We Do after the COVID-19 Vaccination? Vaccine-Associated Diseases and Precautionary Measures against Adverse Reactions.Vaccines (Basel). 2022 May 28;10(6):866. doi: 10.3390/vaccines10060866. Vaccines (Basel). 2022. PMID: 35746474 Free PMC article. Review.
-
How do the severe acute respiratory coronavirus 2 (SARS-CoV-2) and its variants escape the host protective immunity and mediate pathogenesis?Bull Natl Res Cent. 2022;46(1):255. doi: 10.1186/s42269-022-00945-3. Epub 2022 Oct 12. Bull Natl Res Cent. 2022. PMID: 36254244 Free PMC article. Review.
-
Inflammatory reaction to recently applied red tattoo ink after COVID-19.J Cosmet Dermatol. 2022 Dec;21(12):7237-7239. doi: 10.1111/jocd.15389. Epub 2022 Sep 26. J Cosmet Dermatol. 2022. PMID: 36106520 Free PMC article. No abstract available.
-
Effects of Omicron Infection and Changes in Serum Antibody Response to Wild-Type, Delta, and Omicron After a Booster Dose With BNT163b2 Vaccine in Korean Healthcare Workers.J Korean Med Sci. 2023 Apr 3;38(13):e103. doi: 10.3346/jkms.2023.38.e103. J Korean Med Sci. 2023. PMID: 37012688 Free PMC article.
References
-
- Asaduzzaman SAI, Zakaria A, Kheya IS, Fahad N, Sikandar YB, Noor R. A comparative study between the severe acute respiratory syndrome-coronavirus-2, severe acute respiratory syndrome coronavirus, and the Middle East respiratory syndrome coronavirus. Biomed Biotechnol Res J. 2020;4:S65–74. doi: 10.4103/bbrj.bbrj_99_20. - DOI
-
- Barda N, Dagan N, Cohen C, Hernán MA, Lipsitch M, Kohane IS, Reis BY, Balicer RD. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet. 2021;398(10316):2093–2100. doi: 10.1016/S0140-6736(21)02249-2. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
