Methane-Derived Carbon as a Driver for Cyanobacterial Growth
- PMID: 35432228
- PMCID: PMC9010870
- DOI: 10.3389/fmicb.2022.837198
Methane-Derived Carbon as a Driver for Cyanobacterial Growth
Abstract
Methane, a potent greenhouse gas produced in freshwater ecosystems, can be used by methane-oxidizing bacteria (MOB) and can therefore subsidize the pelagic food web with energy and carbon. Consortia of MOB and photoautotrophs have been described in aquatic ecosystems and MOB can benefit from photoautotrophs which produce oxygen, thereby enhancing CH4 oxidation. Methane oxidation can account for accumulation of inorganic carbon (i.e., CO2) and the release of exometabolites that may both be important factors influencing the structure of phytoplankton communities. The consortium of MOB and phototroph has been mainly studied for methane-removing biotechnologies, but there is still little information on the role of these interactions in freshwater ecosystems especially in the context of cyanobacterial growth and bloom development. We hypothesized that MOB could be an alternative C source to support cyanobacterial growth in freshwater systems. We detected low δ13C values in cyanobacterial blooms (the lowest detected value -59.97‰ for Planktothrix rubescens) what could be the result of the use of methane-derived carbon by cyanobacteria and/or MOB attached to their cells. We further proved the presence of metabolically active MOB on cyanobacterial filaments using the fluorescein isothiocyanate (FITC) based activity assay. The PCR results also proved the presence of the pmoA gene in several non-axenic cultures of cyanobacteria. Finally, experiments comprising the co-culture of the cyanobacterium Aphanizomenon gracile with the methanotroph Methylosinus sporium proved that cyanobacterial growth was significantly improved in the presence of MOB, presumably through utilizing CO2 released by MOB. On the other hand, 13C-CH4 labeled incubations showed the uptake and assimilation of MOB-derived metabolites by the cyanobacterium. We also observed a higher growth of MOB in the presence of cyanobacteria under a higher irradiance regime, then when grown alone, underpinning the bidirectional influence with as of yet unknown environmental consequences.
Keywords: co-culture; cyanobacteria; greenhouse gases; isotopes; lakes; methane; methane oxidation.
Copyright © 2022 Cerbin, Pérez, Rybak, Wejnerowski, Konowalczyk, Helmsing, Naus-Wiezer, Meima-Franke, Pytlak, Raaijmakers, Nowak and Bodelier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
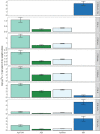
Figures





References
-
- Agasild H., Zingel P., Tuvikene L., Tuvikene A., Timm H., Feldmann T., et al. (2014). Biogenic methane contributes to the food web of a large, shallow lake. Freshw. Biol. 59 272–285. 10.1111/fwb.12263 - DOI
-
- Badr K., Hilliard M., Roberts N., He Q. P., Wang J. (2019). Photoautotroph-methanotroph coculture – a flexible platform for efficient biological CO2-CH4 Co-utilization. IFAC-PapersOnLine 52 916–921. 10.1016/j.ifacol.2019.06.179 - DOI
-
- Balmer M., Downing J. (2011). Carbon dioxide concentrations in eutrophic lakes: undersaturation implies atmospheric uptake. Inland Waters 1 125–132. 10.5268/iw-1.2.366 - DOI
LinkOut - more resources
Full Text Sources

