Unraveling the Effects of Biochemical Drivers on the Bacterial Communities and Volatile Profiles in Refrigerated Sturgeon Filets at 4°C
- PMID: 35432233
- PMCID: PMC9006255
- DOI: 10.3389/fmicb.2022.849236
Unraveling the Effects of Biochemical Drivers on the Bacterial Communities and Volatile Profiles in Refrigerated Sturgeon Filets at 4°C
Abstract
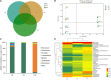
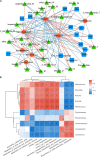
Spoilage bacteria seriously influence the flavor and quality of fish meat. In this study, we investigated the quality characteristics, bacterial community, and volatile profiles of refrigerated (4°C) sturgeon filets during 10-day storage. On day 10, the refrigerated samples showed the lowest bacterial diversity and the largest difference in microbiota and biochemistry. The dominant genera in the fresh samples were Macrococcus, Acinetobacter, Moraxella, Brucella, and Pseudomonas, while the dominant bacteria changed into Acinetobacter, Carnobacterium, Macrococcus, Pseudomonas, and Psychrobacter at the end of storage. Our results suggest that these dominant taxa contribute to the spoilage of the refrigerated sturgeon filets. Meanwhile, during the storage, total viable counts, total volatile basic nitrogen, thiobarbituric acid-reactive substances, and trichloroacetic acid-soluble peptide significantly increased (P < 0.05), while the sensory score decreased steadily. Additionally, the ATP-related compounds and the K-value showed similarly increasing trends. The shelf-life of the refrigerated sturgeon filets was less than 8 days. The gas chromatography-ion mobility spectrometry results suggest that hexanal, ethyl acetate, ethanol, butanal, 1-propanol, isopentyl alcohol, 2-pentanone, 2-heptanone, ethyl propanoate, and propyl sulfide are potential chemical spoilage markers. The predicted metabolic pathways indicated an abundant carbohydrate metabolism and amino metabolism in the refrigerated sturgeon filets. This study provides insight into the determinants of sturgeon shelf-life and the spoilage process involved in refrigerated fish.
Keywords: GC-IMS; high-throughput sequencing; microbial communities; spoilage bacteria; volatile organic compounds.
Copyright © 2022 Tan, Xiao, Wu, Li and Shang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Amerine M. A., Pangborn R. M., Roessler E. B. (1965). Principles of Sensory Evaluation of Food. New York: Academic Press. 10.1016/B978-1-4832-0018-7.50001-5 - DOI
-
- Boziaris I. S., Parlapani F. F. (2017). “Specific spoilage organisms (SSOs) in fish,” in The Microbiological Quality of Food, eds Bevilacqua A., Corbo M. R., Sinigaglia M. (Sawston: Woodhead publishing; ), 61–98. 10.1016/B978-0-08-100502-6.00006-6 - DOI
-
- Chen Y., Cai W., Shi Y., Dong X., Bai F., Shen S., et al. (2020). Effects of different salt concentrations and vacuum packaging on the shelf-stability of Russian sturgeon (Acipenser gueldenstaedti) stored at 4 °C. Food Control. 109:106865. 10.1016/j.foodcont.2019.106865 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

