Identification and Verification of Potential Hub Genes in Amphetamine-Type Stimulant (ATS) and Opioid Dependence by Bioinformatic Analysis
- PMID: 35432486
- PMCID: PMC9006114
- DOI: 10.3389/fgene.2022.837123
Identification and Verification of Potential Hub Genes in Amphetamine-Type Stimulant (ATS) and Opioid Dependence by Bioinformatic Analysis
Abstract
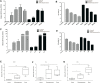
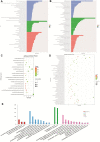
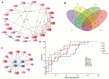
Objective: Amphetamine-type stimulant (ATS) and opioid dependencies are chronic inflammatory diseases with similar symptoms and common genomics. However, their coexpressive genes have not been thoroughly investigated. We aimed to identify and verify the coexpressive hub genes and pathway involved in the pathogenesis of ATS and opioid dependencies. Methods: The microarray of ATS- and opioid-treatment mouse models was obtained from the Gene Expression Omnibus database. GEO2R and Venn diagram were performed to identify differentially expressed genes (DEGs) and coexpressive DEGs (CDEGs). Functional annotation and protein-protein interaction network detected the potential functions. The hub genes were screened using the CytoHubba and MCODE plugin with different algorithms, and further validated by receiver operating characteristic analysis in the GSE15774 database. We also validated the hub genes mRNA levels in BV2 cells using qPCR. Result: Forty-four CDEGs were identified between ATS and opioid databases, which were prominently enriched in the PI3K/Akt signaling pathway. The top 10 hub genes were mainly enriched in apoptotic process (CD44, Dusp1, Sgk1, and Hspa1b), neuron differentiation, migration, and proliferation (Nr4a2 and Ddit4), response to external stimulation (Fos and Cdkn1a), and transcriptional regulation (Nr4a2 and Npas4). Receiver operating characteristic (ROC) analysis found that six hub genes (Fos, Dusp1, Sgk1, Ddit4, Cdkn1a, and Nr4a2) have an area under the curve (AUC) of more than 0.70 in GSE15774. The mRNA levels of Fos, Dusp1, Sgk1, Ddit4, Cdkn1a, PI3K, and Akt in BV2 cells and GSE15774 with METH and heroin treatments were higher than those of controls. However, the Nr4a2 mRNA levels increased in BV2 cells and decreased in the bioinformatic analysis. Conclusions: The identification of hub genes was associated with ATS and opioid dependencies, which were involved in apoptosis, neuron differentiation, migration, and proliferation. The PI3K/Akt signaling pathway might play a critical role in the pathogenesis of substance dependence.
Keywords: PI3K/Akt pathway; amphetamine-type stimulants (ATS); apoptosis; differentially expressed genes (DEGs); hub gene; opioids.
Copyright © 2022 Zhang, Deng, Liu, Ke, Xiang, Ma, Zhang, Yang, Liu and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of Tissue-Specific Expressed Hub Genes and Potential Drugs in Rheumatoid Arthritis Using Bioinformatics Analysis.Front Genet. 2022 Mar 18;13:855557. doi: 10.3389/fgene.2022.855557. eCollection 2022. Front Genet. 2022. PMID: 35368701 Free PMC article.
-
Identification of six candidate genes for endometrial carcinoma by bioinformatics analysis.World J Surg Oncol. 2020 Jul 8;18(1):161. doi: 10.1186/s12957-020-01920-w. World J Surg Oncol. 2020. PMID: 32641130 Free PMC article.
-
Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis.PeerJ. 2018 Jul 2;6:e5180. doi: 10.7717/peerj.5180. eCollection 2018. PeerJ. 2018. PMID: 30002985 Free PMC article.
-
Bioinformatic Analysis Combined With Experimental Validation Reveals Novel Hub Genes and Pathways Associated With Focal Segmental Glomerulosclerosis.Front Mol Biosci. 2022 Jan 4;8:691966. doi: 10.3389/fmolb.2021.691966. eCollection 2021. Front Mol Biosci. 2022. PMID: 35059432 Free PMC article.
-
Functional modular networks identify the pivotal genes associated with morphine addiction and potential drug therapies.BMC Anesthesiol. 2023 May 3;23(1):151. doi: 10.1186/s12871-023-02111-2. BMC Anesthesiol. 2023. Retraction in: BMC Anesthesiol. 2024 Mar 9;24(1):97. doi: 10.1186/s12871-024-02482-0. PMID: 37138216 Free PMC article. Retracted.
References
-
- Bandettini W. P., Kellman P., Mancini C., Booker O. J., Vasu S., Leung S. W., et al. (2012). MultiContrast Delayed Enhancement (MCODE) Improves Detection of Subendocardial Myocardial Infarction by Late Gadolinium Enhancement Cardiovascular Magnetic Resonance: a Clinical Validation Study. J. Cardiovasc. Magn. Reson. 14, 83. 10.1186/1532-429X-14-83 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

