Protective Prognostic Biomarkers Negatively Correlated with Macrophage M2 Infiltration in Low-Grade Glioma
- PMID: 35432538
- PMCID: PMC9012619
- DOI: 10.1155/2022/3623591
Protective Prognostic Biomarkers Negatively Correlated with Macrophage M2 Infiltration in Low-Grade Glioma
Abstract
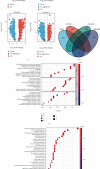
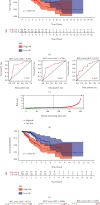
Tumor-associated Macrophages (TAMs) play a vital role in the progression of glioma. Macrophage M2 has been confirmed to promote immunosuppression and proliferation of low-grade glioma (LGG). Here, we searched for genes negatively correlated with Macrophages M2 by bioinformatical methods and investigated their protective ability for prognosis. LGG and adjacent normal samples were screened out in TCGA and three GEO datasets. 326 overlapped differentially expressed genes were calculated, and their biological functions were investigated by Go and KEGG analyses. Macrophage M2 accounted for the highest proportion among all 22 immune cells by CIBERSORT deconvolution algorithm. The proportion of Macrophage M2 in LGG was also higher than that in normal tissue according to several deconvolution algorithms. 43 genes in the blue module negatively correlated with Macrophage M2 infiltration were identified by weighted gene coexpression network analysis (WGCNA). Through immune infiltration and correlation analysis, FGFBP3, VAX2, and SHD were selected and they were enriched in G protein-coupled receptors' signaling regulation and cytokine receptor interaction. They could prolong the overall and disease-free survival time. Univariate and multivariate Cox regression analyses were applied to evaluate prognosis prediction ability. Interestingly, FGFBP3 and AHD were independent prognostic predictors. A nomogram was drawn, and its 1-year, 3-year, and 5-year survival prognostic value was verified by ROC curves and calibration plots. In conclusion, FGFBP3, VAX2, and SHD were protective prognostic biomarkers against Macrophage M2 infiltration in low-grade glioma. The FGFBP3 and SHD were independent factors to effectively predict long-term survival probability.
Copyright © 2022 Yunyang Zhu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures








References
-
- Delgado-López P. D., Corrales-García E. M., Martino J., Lastra-Aras E., Dueñas-Polo M. T. Diffuse low-grade glioma: a review on the new molecular classification, natural history and current management strategies. Clinical & Translational Oncology . 2017;19(8):931–944. doi: 10.1007/s12094-017-1631-4. - DOI - PubMed
LinkOut - more resources
Full Text Sources

