Structure-mechanics statistical learning uncovers mechanical relay in proteins
- PMID: 35432911
- PMCID: PMC8966636
- DOI: 10.1039/d1sc06184d
Structure-mechanics statistical learning uncovers mechanical relay in proteins
Abstract
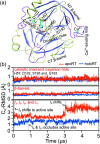
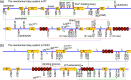
A protein's adaptive response to its substrates is one of the key questions driving molecular physics and physical chemistry. This work employs the recently developed structure-mechanics statistical learning method to establish a mechanical perspective. Specifically, by mapping all-atom molecular dynamics simulations onto the spring parameters of a backbone-side-chain elastic network model, the chemical moiety specific force constants (or mechanical rigidity) are used to assemble the rigidity graph, which is the matrix of inter-residue coupling strength. Using the S1A protease and the PDZ3 signaling domain as examples, chains of spatially contiguous residues are found to exhibit prominent changes in their mechanical rigidity upon substrate binding or dissociation. Such a mechanical-relay picture thus provides a mechanistic underpinning for conformational changes, long-range communication, and inter-domain allostery in both proteins, where the responsive mechanical hotspots are mostly residues having important biological functions or significant mutation sensitivity.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
Statistical learning of protein elastic network from positional covariance matrix.Comput Struct Biotechnol J. 2023 Mar 28;21:2524-2535. doi: 10.1016/j.csbj.2023.03.033. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37095762 Free PMC article.
-
Mechanical couplings of protein backbone and side chains exhibit scale-free network properties and specific hotspots for function.Comput Struct Biotechnol J. 2021 Sep 8;19:5309-5320. doi: 10.1016/j.csbj.2021.09.004. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34765086 Free PMC article.
-
Mechanical variations in proteins with large-scale motions highlight the formation of structural locks.J Struct Biol. 2018 Sep;203(3):195-204. doi: 10.1016/j.jsb.2018.05.006. Epub 2018 May 28. J Struct Biol. 2018. PMID: 29852221
-
Bridging Enzymatic Structure Function via Mechanics: A Coarse-Grain Approach.Methods Enzymol. 2016;578:227-48. doi: 10.1016/bs.mie.2016.05.022. Epub 2016 Jun 9. Methods Enzymol. 2016. PMID: 27497169 Review.
-
Detecting Functional Dynamics in Proteins with Comparative Perturbed-Ensembles Analysis.Acc Chem Res. 2019 Dec 17;52(12):3455-3464. doi: 10.1021/acs.accounts.9b00485. Epub 2019 Dec 3. Acc Chem Res. 2019. PMID: 31793290 Review.
Cited by
-
CHARMM at 45: Enhancements in Accessibility, Functionality, and Speed.J Phys Chem B. 2024 Oct 17;128(41):9976-10042. doi: 10.1021/acs.jpcb.4c04100. Epub 2024 Sep 20. J Phys Chem B. 2024. PMID: 39303207 Free PMC article. Review.
-
Statistical learning of protein elastic network from positional covariance matrix.Comput Struct Biotechnol J. 2023 Mar 28;21:2524-2535. doi: 10.1016/j.csbj.2023.03.033. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37095762 Free PMC article.
-
Mechanical codes of chemical-scale specificity in DNA motifs.Chem Sci. 2023 Aug 29;14(37):10155-10166. doi: 10.1039/d3sc01671d. eCollection 2023 Sep 27. Chem Sci. 2023. PMID: 37772098 Free PMC article.
References
LinkOut - more resources
Full Text Sources

