SuRVoS 2: Accelerating Annotation and Segmentation for Large Volumetric Bioimage Workflows Across Modalities and Scales
- PMID: 35433703
- PMCID: PMC9011330
- DOI: 10.3389/fcell.2022.842342
SuRVoS 2: Accelerating Annotation and Segmentation for Large Volumetric Bioimage Workflows Across Modalities and Scales
Abstract
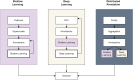
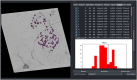
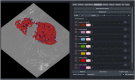
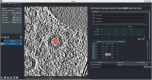
As sample preparation and imaging techniques have expanded and improved to include a variety of options for larger sized and numbers of samples, the bottleneck in volumetric imaging is now data analysis. Annotation and segmentation are both common, yet difficult, data analysis tasks which are required to bring meaning to the volumetric data. The SuRVoS application has been updated and redesigned to provide access to both manual and machine learning-based segmentation and annotation techniques, including support for crowd sourced data. Combining adjacent, similar voxels (supervoxels) provides a mechanism for speeding up segmentation both in the painting of annotation and by training a segmentation model on a small amount of annotation. The support for layers allows multiple datasets to be viewed and annotated together which, for example, enables the use of correlative data (e.g. crowd-sourced annotations or secondary imaging techniques) to guide segmentation. The ability to work with larger data on high-performance servers with GPUs has been added through a client-server architecture and the Pytorch-based image processing and segmentation server is flexible and extensible, and allows the implementation of deep learning-based segmentation modules. The client side has been built around Napari allowing integration of SuRVoS into an ecosystem for open-source image analysis while the server side has been built with cloud computing and extensibility through plugins in mind. Together these improvements to SuRVoS provide a platform for accelerating the annotation and segmentation of volumetric and correlative imaging data across modalities and scales.
Keywords: U-net; X-ray microscopy imaging; annotation; computer vision; open source software; python (programming language); segmentation (image processing); volume electron microscopy (vEM).
Copyright © 2022 Pennington, King, Tun, Ho, Luengo, Darrow and Basham.
Conflict of interest statement
Authors AP, OK, WT, IL, and MB were employed by the company Diamond Light Source Ltd. EH was employed by the Rosalind Franklin Institute. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Abràmoff M. D., Magalhães P. J., Ram S. J. (2004). Image Processing with ImageJ. Biophotonics Int. 11 (7), 36–42.
-
- Alvarez-Borges F. J., King O. N. F., Madhusudhan B. N., Connolley T., Basham M., Ahmed S. I. (2021). U-Net Segmentation Methods for Variable-Contrast XCT Images of Methane-Bearing Sand. Earth and Space Science Open Archive. 10.1002/essoar.10506807.1 - DOI
-
- Basham (2021). GitHub - DiamondLightSource/SuRVoS2: Next Generation of SuRVoS. [online] Available at: https://github.com/DiamondLightSource/SuRVoS2 (Accessed December 13, 2021).
LinkOut - more resources
Full Text Sources
Research Materials

