Characteristics of hepatitis C virus resistance in an international cohort after a decade of direct-acting antivirals
- PMID: 35434589
- PMCID: PMC9010635
- DOI: 10.1016/j.jhepr.2022.100462
Characteristics of hepatitis C virus resistance in an international cohort after a decade of direct-acting antivirals
Abstract
Background & aims: Direct-acting antiviral (DAA) regimens provide a cure in >95% of patients with chronic HCV infection. However, in some patients in whom therapy fails, resistance-associated substitutions (RASs) can develop, limiting retreatment options and risking onward resistant virus transmission. In this study, we evaluated RAS prevalence and distribution, including novel NS5A RASs and clinical factors associated with RAS selection, among patients who experienced DAA treatment failure.
Methods: SHARED is an international consortium of clinicians and scientists studying HCV drug resistance. HCV sequence linked metadata from 3,355 patients were collected from 22 countries. NS3, NS5A, and NS5B RASs in virologic failures, including novel NS5A substitutions, were examined. Associations of clinical and demographic characteristics with RAS selection were investigated.
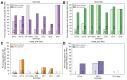
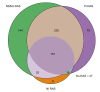
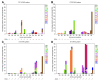
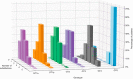
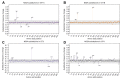
Results: The frequency of RASs increased from its natural prevalence following DAA exposure: 37% to 60% in NS3, 29% to 80% in NS5A, 15% to 22% in NS5B for sofosbuvir, and 24% to 37% in NS5B for dasabuvir. Among 730 virologic failures, most were treated with first-generation DAAs, 94% had drug resistance in ≥1 DAA class: 31% single-class resistance, 42% dual-class resistance (predominantly against protease and NS5A inhibitors), and 21% triple-class resistance. Distinct patterns containing ≥2 highly resistant RASs were common. New potential NS5A RASs and adaptive changes were identified in genotypes 1a, 3, and 4. Following DAA failure, RAS selection was more frequent in older people with cirrhosis and those infected with genotypes 1b and 4.
Conclusions: Drug resistance in HCV is frequent after DAA treatment failure. Previously unrecognized substitutions continue to emerge and remain uncharacterized.
Lay summary: Although direct-acting antiviral medications effectively cure hepatitis C in most patients, sometimes treatment selects for resistant viruses, causing antiviral drugs to be either ineffective or only partially effective. Multidrug resistance is common in patients for whom DAA treatment fails. Older patients and patients with advanced liver diseases are more likely to select drug-resistant viruses. Collective efforts from international communities and governments are needed to develop an optimal approach to managing drug resistance and preventing the transmission of resistant viruses.
Keywords: DAA; DAA, direct-acting antiviral; DCV, daclatasvir; DSV, dasabuvir; GT, genotype; HCV; LDV, ledipasvir; NI, nucleoside; NNI, non-nucleoside; NS5A; NS5AI, NS5A replication complex inhibitor; OR, odds ratio; PI, NS3 protease inhibitor; PIB, pibrentasvir; RAS; RASs, resistance-associated substitutions; SHARED, The Surveillance of Hepatitis C Antiviral Resistance, Epidemiology and methoDologies; SOF, sofosbuvir; SVR, sustained virologic response; VEL, velpatasvir; aOR, adjusted odds ratio; sFC, substitution frequency change; virologic failure.
© 2022 The Authors.
Conflict of interest statement
J.M.P. has been an advisor and/or speaker for AbbVie, Assembly Biosciences, Arbutus, Merck, Gilead, Regulus, and Memo Therapeutics. J.D. receives research support from Gilead. A.Y.M.H. is a consultant for Boston Pharmaceuticals. Outside the submitted work, J.G. reports grants and personal fees from AbbVie, Gilead Sciences, Merck, and Cepheid and grants from Hologic and Indivior. M.W.D has been an advisor and/or speaker for Gilead, AbbVie, and Merck and has received grants from Gilead and AbbVie. F.G.G. has been an advisor and/or speaker for AbbVie, Merck, and Gilead. S.F. has been an advisor and/or speaker for Abbott diagnostics, AbbVie, Gilead, and AB Science. F.C.S. has been an advisor and/or speaker for AbbVie, Merck, and Gilead and received grants from Merck and Gilead. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures

References
-
- Pawlotsky J.-M., Negro F., Aghemo A., Berenguer M., Dalgard O., Dusheiko G., et al. EASL recommendations on treatment of hepatitis C: final update of the series. J Hepatol. 2020;73:1170–1218. - PubMed
-
- Kåberg M., Weiland O. Hepatitis C elimination - macro-elimination. Liver Int. 2020;40(Suppl 1):61–66. - PubMed
-
- International Committee on Taxonomy of Viruses - HCV Classification . 2017. HCV Classification.https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-cl... In.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

