Identification of a Crosstalk among TGR5, GLIS2, and TP53 Signaling Pathways in the Control of Undifferentiated Germ Cell Homeostasis and Chemoresistance
- PMID: 35435331
- PMCID: PMC9189661
- DOI: 10.1002/advs.202200626
Identification of a Crosstalk among TGR5, GLIS2, and TP53 Signaling Pathways in the Control of Undifferentiated Germ Cell Homeostasis and Chemoresistance
Abstract
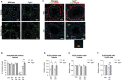
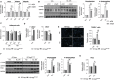
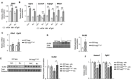
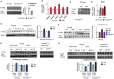
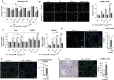
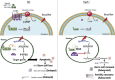
Spermatogonial stem cells regenerate and maintain spermatogenesis throughout life, making testis a good model for studying stem cell biology. The effects of chemotherapy on fertility have been well-documented previously. This study investigates how busulfan, an alkylating agent that is often used for chemotherapeutic purposes, affects male fertility. Specifically, the role of the TGR5 pathway is investigated on spermatogonia homeostasis using in vivo, in vitro, and pharmacological methods. In vivo studies are performed using wild-type and Tgr5-deficient mouse models. The results clearly show that Tgr5 deficiency can facilitate restoration of the spermatogonia homeostasis and allow faster resurgence of germ cell lineage after exposure to busulfan. TGR5 modulates the expression of key genes of undifferentiated spermatogonia such as Gfra1 and Fgfr2. At the molecular level, the present data highlight molecular mechanisms underlying the interactions among the TGR5, GLIS2, and TP53 pathways in spermatogonia associated with germ cell apoptosis following busulfan exposure. This study makes a significant contribution to the literature because it shows that TGR5 plays key role on undifferentiated germ cell homeostasis and that modulating the TGR5 signaling pathway could be used as a potential therapeutic tool for fertility disorders.
Keywords: GLIS2; TGR5; TP53; chemodrugs; germ cells; male fertility; stem cell regeneration.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- de Rooij D. G., Russell L. D., J. Androl. 2000, 21, 776. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
