Identification of environmental chemicals that activate p53 signaling after in vitro metabolic activation
- PMID: 35435491
- PMCID: PMC9151520
- DOI: 10.1007/s00204-022-03291-5
Identification of environmental chemicals that activate p53 signaling after in vitro metabolic activation
Abstract
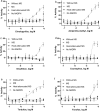
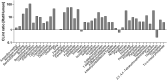
Currently, approximately 80,000 chemicals are used in commerce. Most have little-to-no toxicity information. The U.S. Toxicology in the 21st Century (Tox21) program has conducted a battery of in vitro assays using a quantitative high-throughput screening (qHTS) platform to gain toxicity information on environmental chemicals. Due to technical challenges, standard methods for providing xenobiotic metabolism could not be applied to qHTS assays. To address this limitation, we screened the Tox21 10,000-compound (10K) library, with concentrations ranging from 2.8 nM to 92 µM, using a p53 beta-lactamase reporter gene assay (p53-bla) alone or with rat liver microsomes (RLM) or human liver microsomes (HLM) supplemented with NADPH, to identify compounds that induce p53 signaling after biotransformation. Two hundred and seventy-eight compounds were identified as active under any of these three conditions. Of these 278 compounds, 73 gave more potent responses in the p53-bla assay with RLM, and 2 were more potent in the p53-bla assay with HLM compared with the responses they generated in the p53-bla assay without microsomes. To confirm the role of metabolism in the differential responses, we re-tested these 75 compounds in the absence of NADPH or with heat-attenuated microsomes. Forty-four compounds treated with RLM, but none with HLM, became less potent under these conditions, confirming the role of RLM in metabolic activation. Further evidence of biotransformation was obtained by measuring the half-life of the parent compounds in the presence of microsomes. Together, the data support the use of RLM in qHTS for identifying chemicals requiring biotransformation to induce biological responses.
Keywords: High-throughput metabolism assay; High-throughput screening; In silico metabolism prediction; In vitro metabolism; Microsomes; p53.
© 2022. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

