Microwell Fluoride Screen for Chemical, Enzymatic, and Cellular Reactions Reveals Latent Microbial Defluorination Capacity for -CF3 Groups
- PMID: 35435713
- PMCID: PMC9088286
- DOI: 10.1128/aem.00288-22
Microwell Fluoride Screen for Chemical, Enzymatic, and Cellular Reactions Reveals Latent Microbial Defluorination Capacity for -CF3 Groups
Abstract
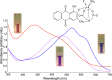
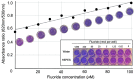
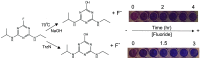
The capacity to defluorinate polyfluorinated organic compounds is a rare phenotype in microbes but is increasingly considered important for maintaining the environment. New discoveries will be greatly facilitated by the ability to screen many natural and engineered microbes in a combinatorial manner against large numbers of fluorinated compounds simultaneously. Here, we describe a low-volume, high-throughput screening method to determine defluorination capacity of microbes and their enzymes. The method is based on selective binding of fluoride to a lanthanum chelate complex that gives a purple-colored product. It was miniaturized to determine biodefluorination in 96-well microtiter plates by visual inspection or robotic handling and spectrophotometry. Chemicals commonly used in microbiological studies were examined to define usable buffers and reagents. Base-catalyzed, purified enzyme and whole-cell defluorination reactions were demonstrated with fluoroatrazine and showed correspondence between the microtiter assay and a fluoride electrode. For discovering new defluorination reactions and mechanisms, a chemical library of 63 fluorinated compounds was screened in vivo with Pseudomonas putida F1 in microtiter well plates. These data were also calibrated against a fluoride electrode. Our new method revealed 21 new compounds undergoing defluorination. A compound with four fluorine substituents, 4-fluorobenzotrifluoride, was shown to undergo defluorination to the greatest extent. The mechanism of its defluorination was studied to reveal a latent microbial propensity to defluorinate trifluoromethylphenyl groups, a moiety that is commonly incorporated into numerous pharmaceutical and agricultural chemicals. IMPORTANCE Thousands of organofluorine chemicals are known, and a number are considered to be persistent and toxic environmental pollutants. Environmental bioremediation methods are avidly being sought, but few bacteria biodegrade fluorinated chemicals. To find new organofluoride biodegradation, a rapid screening method was developed. The method is versatile, monitoring chemical, enzymatic, and whole-cell biodegradation. Biodegradation of organofluorine compounds invariably releases fluoride anions, which was sensitively detected. Our method uncovered 21 new microbial defluorination reactions. A general mechanism was delineated for the biodegradation of trifluoromethylphenyl groups that are increasingly being used in drugs and pesticides.
Keywords: PFAS; Pseudomonas putida F1; bacteria; defluorination; fluoride; high throughput; organofluorine; screening; trifluoromethyl.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Unexpected Mechanism of Biodegradation and Defluorination of 2,2-Difluoro-1,3-Benzodioxole by Pseudomonas putida F1.mBio. 2021 Dec 21;12(6):e0300121. doi: 10.1128/mBio.03001-21. Epub 2021 Nov 16. mBio. 2021. PMID: 34781746 Free PMC article.
-
Microwell fluoride assay screening for enzymatic defluorination.Methods Enzymol. 2024;696:65-83. doi: 10.1016/bs.mie.2023.12.020. Epub 2024 Jan 11. Methods Enzymol. 2024. PMID: 38658089
-
Why Is the Biodegradation of Polyfluorinated Compounds So Rare?mSphere. 2021 Oct 27;6(5):e0072121. doi: 10.1128/mSphere.00721-21. Epub 2021 Oct 13. mSphere. 2021. PMID: 34643420 Free PMC article.
-
The link between ancient microbial fluoride resistance mechanisms and bioengineering organofluorine degradation or synthesis.Nat Commun. 2024 May 30;15(1):4593. doi: 10.1038/s41467-024-49018-1. Nat Commun. 2024. PMID: 38816380 Free PMC article. Review.
-
Confronting PFAS persistence: enzymes catalyzing C-F bond cleavage.Trends Biochem Sci. 2025 Jan;50(1):71-83. doi: 10.1016/j.tibs.2024.11.001. Epub 2024 Dec 5. Trends Biochem Sci. 2025. PMID: 39643519 Review.
Cited by
-
Evolutionary obstacles and not C-F bond strength make PFAS persistent.Microb Biotechnol. 2024 Apr;17(4):e14463. doi: 10.1111/1751-7915.14463. Microb Biotechnol. 2024. PMID: 38593328 Free PMC article.
-
Enzymatic carbon-fluorine bond cleavage by human gut microbes.Proc Natl Acad Sci U S A. 2025 Jun 17;122(24):e2504122122. doi: 10.1073/pnas.2504122122. Epub 2025 Jun 13. Proc Natl Acad Sci U S A. 2025. PMID: 40512801 Free PMC article.
-
Strategies for the Biodegradation of Polyfluorinated Compounds.Microorganisms. 2022 Aug 17;10(8):1664. doi: 10.3390/microorganisms10081664. Microorganisms. 2022. PMID: 36014082 Free PMC article. Review.
-
Fluoro-recognition: New in vivo fluorescent assay for toluene dioxygenase probing induction by and metabolism of polyfluorinated compounds.Environ Microbiol. 2022 Nov;24(11):5202-5216. doi: 10.1111/1462-2920.16187. Epub 2022 Oct 17. Environ Microbiol. 2022. PMID: 36054238 Free PMC article.
-
A prescription for engineering PFAS biodegradation.Biochem J. 2024 Dec 4;481(23):1757-1770. doi: 10.1042/BCJ20240283. Biochem J. 2024. PMID: 39585294 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

