Class C β-Lactamases: Molecular Characteristics
- PMID: 35435729
- PMCID: PMC9491196
- DOI: 10.1128/cmr.00150-21
Class C β-Lactamases: Molecular Characteristics
Abstract
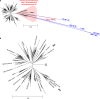
Class C β-lactamases or cephalosporinases can be classified into two functional groups (1, 1e) with considerable molecular variability (≤20% sequence identity). These enzymes are mostly encoded by chromosomal and inducible genes and are widespread among bacteria, including Proteobacteria in particular. Molecular identification is based principally on three catalytic motifs (64SXSK, 150YXN, 315KTG), but more than 70 conserved amino-acid residues (≥90%) have been identified, many close to these catalytic motifs. Nevertheless, the identification of a tiny, phylogenetically distant cluster (including enzymes from the genera Legionella, Bradyrhizobium, and Parachlamydia) has raised questions about the possible existence of a C2 subclass of β-lactamases, previously identified as serine hydrolases. In a context of the clinical emergence of extended-spectrum AmpC β-lactamases (ESACs), the genetic modifications observed in vivo and in vitro (point mutations, insertions, or deletions) during the evolution of these enzymes have mostly involved the Ω- and H-10/R2-loops, which vary considerably between genera, and, in some cases, the conserved triplet 150YXN. Furthermore, the conserved deletion of several amino-acid residues in opportunistic pathogenic species of Acinetobacter, such as A. baumannii, A. calcoaceticus, A. pittii and A. nosocomialis (deletion of residues 304-306), and in Hafnia alvei and H. paralvei (deletion of residues 289-290), provides support for the notion of natural ESACs. The emergence of higher levels of resistance to β-lactams, including carbapenems, and to inhibitors such as avibactam is a reality, as the enzymes responsible are subject to complex regulation encompassing several other genes (ampR, ampD, ampG, etc.). Combinations of resistance mechanisms may therefore be at work, including overproduction or change in permeability, with the loss of porins and/or activation of efflux systems.
Keywords: AmpC β-lactamases; ESAC; cephalosporinases; extended-spectrum; phylogeny; primary structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
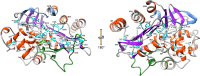

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

