Laser Microdissection for Species-Agnostic Single-Tissue Applications
- PMID: 35435919
- PMCID: PMC9976942
- DOI: 10.3791/63666
Laser Microdissection for Species-Agnostic Single-Tissue Applications
Abstract
Single-cell methodologies have revolutionized the analysis of the transcriptomes of specific cell types. However, they often require species-specific genetic "toolkits," such as promoters driving tissue-specific expression of fluorescent proteins. Further, protocols that disrupt tissues to isolate individual cells remove cells from their native environment (e.g., signaling from neighbors) and may result in stress responses or other differences from native gene expression states. In the present protocol, laser microdissection (LMD) is optimized to isolate individual nematode tail tips for the study of gene expression during male tail tip morphogenesis. LMD allows the isolation of a portion of the animal without the need for cellular disruption or species-specific toolkits and is thus applicable to any species. Subsequently, single-cell RNA-seq library preparation protocols such as CEL-Seq2 can be applied to LMD-isolated single tissues and analyzed using standard pipelines, given that a well-annotated genome or transcriptome is available for the species. Such data can be used to establish how conserved or different the transcriptomes are that underlie the development of that tissue in different species. Limitations include the ability to cut out the tissue of interest and the sample size. A power analysis shows that as few as 70 tail tips per condition are required for 80% power. Tight synchronization of development is needed to obtain this number of animals at the same developmental stage. Thus, a method to synchronize animals at 1 h intervals is also described.
Conflict of interest statement
Disclosures
All authors declare that they have no conflicts of interest.
Figures
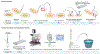


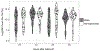

References
-
- Sommer RJ, Bumbarger DJ Nematode model systems in evolution and development. Wiley Interdisciplinary Reviews: Developmental Biology. 1 (3), 389–400 (2012). - PubMed
-
- WormBase. https://wormbase.org/#012-3-6 (2022).
-
- WormAtlas. https://www.wormatlas.org (2022).
-
- WormBook. http://wormbook.org (2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
