The ERK2-DBP domain opposes pathogenesis of a mouse JAK2V617F-driven myeloproliferative neoplasm
- PMID: 35436326
- PMCID: PMC9335498
- DOI: 10.1182/blood.2021013068
The ERK2-DBP domain opposes pathogenesis of a mouse JAK2V617F-driven myeloproliferative neoplasm
Abstract
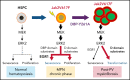
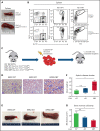
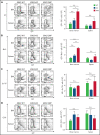
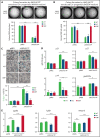
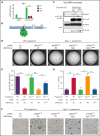
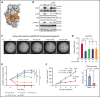
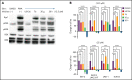
Although Ras/mitogen-activated protein kinase (MAPK) signaling is activated in most human cancers, attempts to target this pathway using kinase-active site inhibitors have not typically led to durable clinical benefit. To address this shortcoming, we sought to test the feasibility of an alternative targeting strategy, focused on the ERK2 substrate binding domains, D and DEF binding pocket (DBP). Disabling the ERK2-DBP domain in mice caused baseline erythrocytosis. Consequently, we investigated the role of the ERK2-D and -DBP domains in disease, using a JAK2-dependent model of polycythemia vera (PV). Of note, inactivation of the ERK2-DBP domain promoted the progression of disease from PV to myelofibrosis, suggesting that the ERK2-DBP domain normally opposes progression. ERK2-DBP inactivation also prevented oncogenic JAK2 kinase (JAK2V617F) from promoting oncogene-induced senescence in vitro. The ERK2-DBP mutation attenuated JAK2-mediated oncogene-induced senescence by preventing the physical interaction of ERK2 with the transcription factor Egr1. Because inactivation of the ERK2-DBP created a functional ERK2 kinase limited to binding substrates through its D domain, these data suggested that the D domain substrates were responsible for promoting oncogene-induced progenitor growth and tumor progression and that pharmacologic targeting of the ERK2-D domain may attenuate cancer cell growth. Indeed, pharmacologic agents targeting the ERK2-D domain were effective in attenuating the growth of JAK2-dependent myeloproliferative neoplasm cell lines. Taken together, these data indicate that the ERK-D and -DBP domains can play distinct roles in the progression of neoplasms and that the D domain has the potential to be a potent therapeutic target in Ras/MAPK-dependent cancers.
© 2022 by The American Society of Hematology.
Figures

Comment in
-
The 2 faces of ERK2 in MPNs.Blood. 2022 Jul 28;140(4):298-300. doi: 10.1182/blood.2022016536. Blood. 2022. PMID: 35900786 No abstract available.
References
-
- Klomp JE, Klomp JA, Der CJ. The ERK mitogen-activated protein kinase signaling network: The final frontier in RAS signal transduction. Biochem Soc Trans. 2021; 49(1):253-267. - PubMed
-
- Korzeniecki C, Priefer R. Targeting KRAS mutant cancers by preventing signaling transduction in the MAPK pathway. Eur J Med Chem. 2021;211:113006. - PubMed
-
- Murphy LO, Blenis J. MAPK signal specificity: The right place at the right time. Trends Biochem Sci. 2006;31(5):268-275. - PubMed
-
- Lu Z, Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 2006; 58(11):621-631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

