Fast and Effective Prediction of the Absolute Binding Free Energies of Covalent Inhibitors of SARS-CoV-2 Main Protease and 20S Proteasome
- PMID: 35436404
- PMCID: PMC9359807
- DOI: 10.1021/jacs.2c00853
Fast and Effective Prediction of the Absolute Binding Free Energies of Covalent Inhibitors of SARS-CoV-2 Main Protease and 20S Proteasome
Abstract
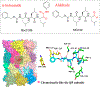
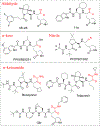
The COVID-19 pandemic has been a public health emergency with continuously evolving deadly variants around the globe. Among many preventive and therapeutic strategies, the design of covalent inhibitors targeting the main protease (Mpro) of SARS-CoV-2 that causes COVID-19 has been one of the hotly pursued areas. Currently, about 30% of marketed drugs that target enzymes are covalent inhibitors. Such inhibitors have been shown in recent years to have many advantages that counteract past reservation of their potential off-target activities, which can be minimized by modulation of the electrophilic warhead and simultaneous optimization of nearby noncovalent interactions. This process can be greatly accelerated by exploration of binding affinities using computational models, which are not well-established yet due to the requirement of capturing the chemical nature of covalent bond formation. Here, we present a robust computational method for effective prediction of absolute binding free energies (ABFEs) of covalent inhibitors. This is done by integrating the protein dipoles Langevin dipoles method (in the PDLD/S-LRA/β version) with quantum mechanical calculations of the energetics of the reaction of the warhead and its amino acid target, in water. This approach evaluates the combined effects of the covalent and noncovalent contributions. The applicability of the method is illustrated by predicting the ABFEs of covalent inhibitors of SARS-CoV-2 Mpro and the 20S proteasome. Our results are found to be reliable in predicting ABFEs for cases where the warheads are significantly different. This computational protocol might be a powerful tool for designing effective covalent inhibitors especially for SARS-CoV-2 Mpro and for targeted protein degradation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

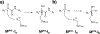

Similar articles
-
In Silico Drug Repositioning to Target the SARS-CoV-2 Main Protease as Covalent Inhibitors Employing a Combined Structure-Based Virtual Screening Strategy of Pharmacophore Models and Covalent Docking.Int J Mol Sci. 2022 Apr 3;23(7):3987. doi: 10.3390/ijms23073987. Int J Mol Sci. 2022. PMID: 35409348 Free PMC article.
-
Estimating the binding energetics of reversible covalent inhibitors of the SARS-CoV-2 main protease: an in silico study.Phys Chem Chem Phys. 2022 Oct 5;24(38):23391-23401. doi: 10.1039/d2cp03080b. Phys Chem Chem Phys. 2022. PMID: 36128834
-
Targeting SARS-CoV-2 Main Protease for Treatment of COVID-19: Covalent Inhibitors Structure-Activity Relationship Insights and Evolution Perspectives.J Med Chem. 2022 Oct 13;65(19):12500-12534. doi: 10.1021/acs.jmedchem.2c01005. Epub 2022 Sep 28. J Med Chem. 2022. PMID: 36169610 Free PMC article. Review.
-
Covalent and non-covalent binding free energy calculations for peptidomimetic inhibitors of SARS-CoV-2 main protease.Phys Chem Chem Phys. 2021 Mar 21;23(11):6746-6757. doi: 10.1039/d1cp00266j. Epub 2021 Mar 12. Phys Chem Chem Phys. 2021. PMID: 33711090
-
Docking Paradigm in Drug Design.Curr Top Med Chem. 2021;21(6):507-546. doi: 10.2174/1568026620666201207095626. Curr Top Med Chem. 2021. PMID: 33292135 Review.
Cited by
-
Elucidation of the α-Ketoamide Inhibition Mechanism: Revealing the Critical Role of the Electrostatic Reorganization Effect of Asp17 in the Active Site of the 20S Proteasome.ACS Catal. 2023 Nov 3;13(21):14368-14376. doi: 10.1021/acscatal.3c03538. Epub 2023 Oct 25. ACS Catal. 2023. PMID: 39188993 Free PMC article.
-
Recent Advances in Alchemical Binding Free Energy Calculations for Drug Discovery.ACS Med Chem Lett. 2023 Feb 16;14(3):244-250. doi: 10.1021/acsmedchemlett.2c00541. eCollection 2023 Mar 9. ACS Med Chem Lett. 2023. PMID: 36923913 Free PMC article. Review.
-
Exploring the Phospholipid Transport Mechanism of ATP8A1-CDC50.Biomedicines. 2023 Feb 13;11(2):546. doi: 10.3390/biomedicines11020546. Biomedicines. 2023. PMID: 36831082 Free PMC article.
-
Perspectives on Computational Enzyme Modeling: From Mechanisms to Design and Drug Development.ACS Omega. 2024 Feb 8;9(7):7393-7412. doi: 10.1021/acsomega.3c09084. eCollection 2024 Feb 20. ACS Omega. 2024. PMID: 38405524 Free PMC article. Review.
-
Energetic and structural insights behind calcium induced conformational transition in calmodulin.Proteins. 2024 Mar;92(3):384-394. doi: 10.1002/prot.26620. Epub 2023 Nov 1. Proteins. 2024. PMID: 37915244 Free PMC article.
References
-
- Schirmeister T; Kesselring J; Jung S; Schneider TH; Weickert A; Becker J; Lee W; Bamberger D; Wich PR; Distler U; Tenzer S; Johé P; Hellmich UA; Engels B Quantum Chemical-Based Protocol for the Rational Design of Covalent Inhibitors. J. Am. Chem. Soc. 2016, 138 (27), 8332–8335. - PubMed
-
- Saha A; Shih AY; Mirzadegan T; Seierstad M Predicting the Binding of Fatty Acid Amide Hydrolase Inhibitors by Free Energy Perturbation. J. Chem. Theory Comput. 2018, 14 (11), 5815–5822. - PubMed
-
- Yu HS; Gao C; Lupyan D; Wu Y; Kimura T; Wu C; Jacobson L; Harder E; Abel R; Wang L Toward Atomistic Modeling of Irreversible Covalent Inhibitor Binding Kinetics. J. Chem. Inf. Model. 2019, 59 (9), 3955–3967. - PubMed
-
- Cournia Z; Allen B; Sherman W Relative Binding Free Energy Calculations in Drug Discovery: Recent Advances and Practical Considerations. J. Chem. Inf. Model. 2017, 57 (12), 2911–2937. - PubMed
-
- Kuhn B; Tichý M; Wang L; Robinson S; Martin RE; Kuglstatter A; Benz J; Giroud M; Schirmeister T; Abel R; Diederich F; Hert J Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors. J. Med. Chem. 2017, 60 (6), 2485–2497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

