Identification of novel prognostic targets in glioblastoma using bioinformatics analysis
- PMID: 35436915
- PMCID: PMC9014588
- DOI: 10.1186/s12938-022-00995-8
Identification of novel prognostic targets in glioblastoma using bioinformatics analysis
Abstract
Background: Glioblastoma (GBM) is the most malignant grade of glioma. Highly aggressive characteristics of GBM and poor prognosis cause GBM-related deaths. The potential prognostic biomarkers remain to be demonstrated. This research builds up predictive gene targets of expression alterations in GBM utilizing bioinformatics analysis.
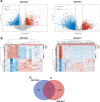
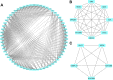
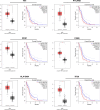
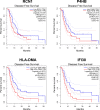
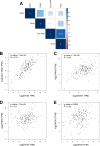
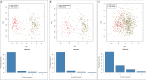
Methods and results: The microarray datasets (GSE15824 and GSE16011) associated with GBM were obtained from Gene Expression Omnibus (GEO) database to identify the differentially expressed genes (DEGs) between GBM and non-tumor tissues. In total, 719 DEGs were obtained and subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for function enrichment analysis. Furthermore, we constructed protein-protein Interaction (PPI) network among DEGs utilizing Search Tool for the Retrieval of Interacting Genes (STRING) online tool and Cytoscape software. The DEGs of degree > 10 was selected as hub genes, including 73 upregulated genes and 21 downregulated genes. Moreover, MCODE application in Cytoscape software was employed to identify three key modules involved in GBM development and prognosis. Additionally, we used the Gene expression profiling and interactive analyses (GEPIA) online tool to further confirm four genes involving in poor prognosis of GBM patients, including interferon-gamma-inducible protein 30 (IFI30), major histocompatibility complex class II-DM alpha (HLA-DMA), Prolyl 4-hydroxylase beta polypeptide (P4HB) and reticulocalbin-1 (RCN1). Furthermore, the correlation analysis indicated that the expression of IFI30, an acknowledged biomarker in glioma, was positively correlated with HLA-DMA, P4HB and RCN1. RCN1 expression was positively correlated with P4HB and HLA-DMA. Moreover, qRT-PCR and immunohistochemistry analysis further validated the upregulation of four prognostic markers in GBM tissues.
Conclusions: Analysis of multiple datasets combined with global network information and experimental verification presents a successful approach to uncover the risk hub genes and prognostic markers of GBM. Our study identified four risk- and prognostic-related gene signatures, including IFI30, HLA-DMA, P4HB and RCN1. This gene sets contribute a new perspective to improve the diagnostic, prognostic, and therapeutic outcomes of GBM.
Keywords: Bioinformatics analysis; Biomarker; Glioblastoma multiform; Prognosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
Identification of potential crucial genes and molecular mechanisms in glioblastoma multiforme by bioinformatics analysis.Mol Med Rep. 2020 Aug;22(2):859-869. doi: 10.3892/mmr.2020.11160. Epub 2020 May 20. Mol Med Rep. 2020. PMID: 32467990 Free PMC article.
-
Multi-omics analysis predicts fibronectin 1 as a prognostic biomarker in glioblastoma multiforme.Genomics. 2022 May;114(3):110378. doi: 10.1016/j.ygeno.2022.110378. Epub 2022 May 2. Genomics. 2022. PMID: 35513291
-
Common gene signatures and key pathways in hypopharyngeal and esophageal squamous cell carcinoma: Evidence from bioinformatic analysis.Medicine (Baltimore). 2020 Oct 16;99(42):e22434. doi: 10.1097/MD.0000000000022434. Medicine (Baltimore). 2020. PMID: 33080677 Free PMC article.
-
mRNA markers for survival prediction in glioblastoma multiforme patients: a systematic review with bioinformatic analyses.BMC Cancer. 2024 May 21;24(1):612. doi: 10.1186/s12885-024-12345-z. BMC Cancer. 2024. PMID: 38773447 Free PMC article.
Cited by
-
A Novel Predictive Model Utilizing Retinal Microstructural Features for Estimating Survival Outcome in Patients with Glioblastoma.Res Sq [Preprint]. 2024 May 17:rs.3.rs-4420925. doi: 10.21203/rs.3.rs-4420925/v1. Res Sq. 2024. Update in: Clin Neurol Neurosurg. 2025 Mar;250:108790. doi: 10.1016/j.clineuro.2025.108790. PMID: 38798600 Free PMC article. Updated. Preprint.
-
Identification of a Prognostic Gene Signature Based on Lipid Metabolism-Related Genes in Esophageal Squamous Cell Carcinoma.Pharmgenomics Pers Med. 2023 Nov 4;16:959-972. doi: 10.2147/PGPM.S430786. eCollection 2023. Pharmgenomics Pers Med. 2023. PMID: 38023824 Free PMC article.
-
A novel predictive model utilizing retinal microstructural features for estimating survival outcome in patients with glioblastoma.Clin Neurol Neurosurg. 2025 Mar;250:108790. doi: 10.1016/j.clineuro.2025.108790. Epub 2025 Feb 17. Clin Neurol Neurosurg. 2025. PMID: 39987704
-
Potential diagnostic markers and therapeutic targets for DM2 and periodontitis based on bioinformatics analysis.PLoS One. 2025 Apr 2;20(4):e0320061. doi: 10.1371/journal.pone.0320061. eCollection 2025. PLoS One. 2025. PMID: 40173189 Free PMC article.
-
Quantitative Evaluation of Stem-like Markers of Human Glioblastoma Using Single-Cell RNA Sequencing Datasets.Cancers (Basel). 2023 Mar 2;15(5):1557. doi: 10.3390/cancers15051557. Cancers (Basel). 2023. PMID: 36900348 Free PMC article.
References
-
- Wirsching HG, Galanis E, Weller M. Glioblastoma. Handb Clin Neurol. 2016;134:381–397. - PubMed
-
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. - PubMed
-
- Hu M, et al. Human cytomegalovirus infection activates glioma activating transcription factor 5 via microRNA in a stress-induced manner. ACS Chem Neurosci. 2021;12(20):3947–3956. - PubMed
-
- Marenco-Hillembrand L, et al. Trends in glioblastoma: outcomes over time and type of intervention: a systematic evidence based analysis. J Neurooncol. 2020;147(2):297–307. - PubMed
-
- Hu M, et al. ELF1 Transcription Factor Enhances the Progression of Glioma via ATF5 promoter. ACS Chem Neurosci. 2021;12(7):1252–1261. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

