LAG3 associates with TCR-CD3 complexes and suppresses signaling by driving co-receptor-Lck dissociation
- PMID: 35437325
- PMCID: PMC9106921
- DOI: 10.1038/s41590-022-01176-4
LAG3 associates with TCR-CD3 complexes and suppresses signaling by driving co-receptor-Lck dissociation
Abstract
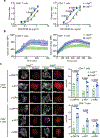
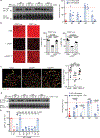
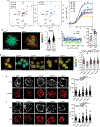
LAG3 is an inhibitory receptor that is highly expressed on exhausted T cells. Although LAG3-targeting immunotherapeutics are currently in clinical trials, how LAG3 inhibits T cell function remains unclear. Here, we show that LAG3 moved to the immunological synapse and associated with the T cell receptor (TCR)-CD3 complex in CD4+ and CD8+ T cells, in the absence of binding to major histocompatibility complex class II-its canonical ligand. Mechanistically, a phylogenetically conserved, acidic, tandem glutamic acid-proline repeat in the LAG3 cytoplasmic tail lowered the pH at the immune synapse and caused dissociation of the tyrosine kinase Lck from the CD4 or CD8 co-receptor, which resulted in a loss of co-receptor-TCR signaling and limited T cell activation. These observations indicated that LAG3 functioned as a signal disruptor in a major histocompatibility complex class II-independent manner, and provide insight into the mechanism of action of LAG3-targeting immunotherapies.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
CONFLICTS OF INTEREST
The authors declare competing financial interests. D.M.M. is an employee and stock holder of Dewpoint Therapeutics. D.A.A.V. and C.J.W. have patents covering LAG3, with others pending, and are entitled to a share in net income generated from licensing of these patent rights for commercial development. DAAV: cofounder and stock holder – Novasenta, Potenza, Tizona, Trishula; stock holder – Oncorus, Werewolf, Apeximmune; patents licensed and royalties - Astellas, BMS, Novasenta; scientific advisory board member - Tizona, Werewolf, F-Star, Bicara, Apeximmune, T7/Imreg Bio; consultant - Astellas, BMS, Almirall, Incyte, G1 Therapeutics; research funding – BMS, Astellas and Novasenta. The remaining authors declare no competing interests.
Figures





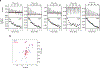




Comment in
-
LAG3 disrupts the TCR signal by local acidification.Nat Immunol. 2022 May;23(5):649-651. doi: 10.1038/s41590-022-01196-0. Nat Immunol. 2022. PMID: 35477982 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

