Bronchial epithelium epithelial-mesenchymal plasticity forms aberrant basaloid-like cells in vitro
- PMID: 35438006
- PMCID: PMC9142163
- DOI: 10.1152/ajplung.00254.2021
Bronchial epithelium epithelial-mesenchymal plasticity forms aberrant basaloid-like cells in vitro
Abstract
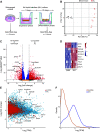
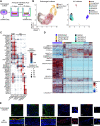
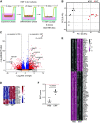
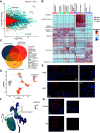
Although epithelial-mesenchymal transition (EMT) is a common feature of fibrotic lung disease, its role in fibrogenesis is controversial. Recently, aberrant basaloid cells were identified in fibrotic lung tissue as a novel epithelial cell type displaying a partial EMT phenotype. The developmental origin of these cells remains unknown. To elucidate the role of EMT in the development of aberrant basaloid cells from the bronchial epithelium, we mapped EMT-induced transcriptional changes at the population and single-cell levels. Human bronchial epithelial cells grown as submerged or air-liquid interface (ALI) cultures with or without EMT induction were analyzed by bulk and single-cell RNA-Sequencing. Comparison of submerged and ALI cultures revealed differential expression of 8,247 protein coding (PC) and 1,621 long noncoding RNA (lncRNA) genes and revealed epithelial cell-type-specific lncRNAs. Similarly, EMT induction in ALI cultures resulted in robust transcriptional reprogramming of 6,020 PC and 907 lncRNA genes. Although there was no evidence for fibroblast/myofibroblast conversion following EMT induction, cells displayed a partial EMT gene signature and an aberrant basaloid-like cell phenotype. The substantial transcriptional differences between submerged and ALI cultures highlight that care must be taken when interpreting data from submerged cultures. This work supports that lung epithelial EMT does not generate fibroblasts/myofibroblasts and confirms ALI cultures provide a physiologically relevant system to study aberrant basaloid-like cells and mechanisms of EMT. We provide a catalog of PC and lncRNA genes and an interactive browser (https://bronc-epi-in-vitro.cells.ucsc.edu/) of single-cell RNA-Seq data for further exploration of potential roles in the lung epithelium in health and lung disease.
Keywords: EMT; ILD; aberrant basaloid cells; epithelial-mesenchymal plasticity (EMP); fibrosis.
Conflict of interest statement
N. Kaminski served as a consultant to Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Theravance, LifeMax, Three Lake Partners, Optikira, Astra Zeneca, RohBar, Veracyte, Augmanity, CSL Behring, Galapagos, Gilead, and Thyron over the past 3 years, reports Equity in Pliant and Thyron, and received a grant from Veracyte, Boehringer Ingelheim, BMS, and nonfinancial support from MiRagen and Astra Zeneca. N. Kaminski has IP on novel biomarkers and therapeutics in IPF licensed to Biotech. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
Figures


References
-
- Nishioka M, Venkatesan N, Dessalle K, Mogas A, Kyoh S, Lin TY, Nair P, Baglole CJ, Eidelman DH, Ludwig MS, Hamid Q. Fibroblast-epithelial cell interactions drive epithelial-mesenchymal transition differently in cells from normal and COPD patients. Respir Res 16: 72, 2015. doi:10.1186/s12931-015-0232-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- figshare/10.6084/m9.figshare.19093763
- figshare/10.6084/m9.figshare.19093730
- figshare/10.6084/m9.figshare.19093736
- figshare/10.6084/m9.figshare.19093724
- figshare/10.6084/m9.figshare.19093754
- figshare/10.6084/m9.figshare.19093727
- figshare/10.6084/m9.figshare.19093745
- figshare/10.6084/m9.figshare.19093742
- figshare/10.6084/m9.figshare.19093733
- figshare/10.6084/m9.figshare.19093751
- figshare/10.6084/m9.figshare.19093739
- figshare/10.6084/m9.figshare.19093757
- figshare/10.6084/m9.figshare.19093760
- figshare/10.6084/m9.figshare.19093748
- figshare/10.6084/m9.figshare.19093703
- figshare/10.6084/m9.figshare.19093700
- figshare/10.6084/m9.figshare.19093709
- figshare/10.6084/m9.figshare.19093712
- figshare/10.6084/m9.figshare.19093706
- figshare/10.6084/m9.figshare.19093721
- figshare/10.6084/m9.figshare.19093718
- figshare/10.6084/m9.figshare.19093715
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

