Bacterial RNA chaperones and chaperone-like riboregulators: behind the scenes of RNA-mediated regulation of cellular metabolism
- PMID: 35438047
- PMCID: PMC9037510
- DOI: 10.1080/15476286.2022.2048565
Bacterial RNA chaperones and chaperone-like riboregulators: behind the scenes of RNA-mediated regulation of cellular metabolism
Abstract
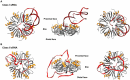
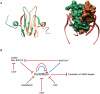
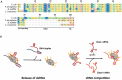
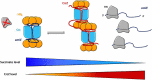
In all domains of life, RNA chaperones safeguard and guide the fate of the cellular RNA pool. RNA chaperones comprise structurally diverse proteins that ensure proper folding, stability, and ribonuclease resistance of RNA, and they support regulatory activities mediated by RNA. RNA chaperones constitute a topologically diverse group of proteins that often present an unstructured region and bind RNA with limited nucleotide sequence preferences. In bacteria, three main proteins - Hfq, ProQ, and CsrA - have been shown to regulate numerous complex processes, including bacterial growth, stress response and virulence. Hfq and ProQ have well-studied activities as global chaperones with pleiotropic impact, while CsrA has a chaperone-like role with more defined riboregulatory function. Here, we describe relevant novel insights into their common features, including RNA binding properties, unstructured domains, and interplay with other proteins important to RNA metabolism.
Keywords: C-terminal domain; CsrA; Hfq; ProQ; RNA chaperone; RNA metabolism; Riboregulation.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Tompa P, Csermely P.. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. - PubMed
-
- Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7(10):834–837. - PubMed
-
- Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8(13):1575–1588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
