PD-1 and ICOS coexpression identifies tumor-reactive CD4+ T cells in human solid tumors
- PMID: 35439168
- PMCID: PMC9197519
- DOI: 10.1172/JCI156821
PD-1 and ICOS coexpression identifies tumor-reactive CD4+ T cells in human solid tumors
Abstract
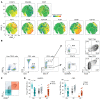
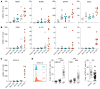
CD4+ Th cells play a key role in orchestrating immune responses, but the identity of the CD4+ Th cells involved in the antitumor immune response remains to be defined. We analyzed the immune cell infiltrates of head and neck squamous cell carcinoma and colorectal cancers and identified a subset of CD4+ Th cells distinct from FOXP3+ Tregs that coexpressed programmed cell death 1 (PD-1) and ICOS. These tumor-infiltrating lymphocyte CD4+ Th cells (CD4+ Th TILs) had a tissue-resident memory phenotype, were present in MHC class II-rich areas, and proliferated in the tumor, suggesting local antigen recognition. The T cell receptor repertoire of the PD-1+ICOS+ CD4+ Th TILs was oligoclonal, with T cell clones expanded in the tumor, but present at low frequencies in the periphery. Finally, these PD-1+ICOS+ CD4+ Th TILs were shown to recognize both tumor-associated antigens and tumor-specific neoantigens. Our findings provide an approach for isolating tumor-reactive CD4+ Th TILs directly ex vivo that will help define their role in the antitumor immune response and potentially improve future adoptive T cell therapy approaches.
Keywords: Cancer; Immunology; Immunotherapy; Oncology; T cells.
Figures






Comment in
-
Finding the right help in the tumor microenvironment.J Clin Invest. 2022 Jun 15;132(12):e161052. doi: 10.1172/JCI161052. J Clin Invest. 2022. PMID: 35703176 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

