UHRF1 establishes crosstalk between somatic and germ cells in male reproduction
- PMID: 35440090
- PMCID: PMC9018721
- DOI: 10.1038/s41419-022-04837-2
UHRF1 establishes crosstalk between somatic and germ cells in male reproduction
Abstract
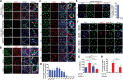
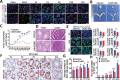
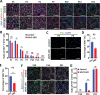
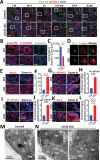
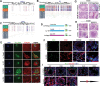
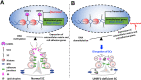
Sertoli cells (SCs) support and nourish germ cells (GCs) through their crosstalk during spermatogenesis. However, the underlying epigenetic mechanism that ensures SCs' functions in this process remains unclear. Here, we report that UHRF1, a critical epigenetic regulator, is mainly expressed in human and mouse pre-mature SCs, and is essential for establishing Sertoli-Germ cell crosstalk. SC-specific UHRF1 knockout mice exhibit complete sterility with Sertoli cell (SC) proliferation and differentiation aberrance, blood-testis barrier (BTB) disruption, and immature germ cell (GC) sloughing. RNA sequencing and Whole Genome Bisulfite Sequencing (WGBS) revealed that many extracellular matrix (ECM)-related genes (e.g., Timp1, Trf, and Spp1) appeared upregulated with the DNA hypomethylation status in UHRF1-deficient SCs. Strikingly, overexpression of Timp1, Trf, and Spp1 in SCs in vitro and in vivo could phenocopy the SC-specific UHRF1-deficient mice. Our data demonstrated that UHRF1 regulates the transcriptional program of ECM-related genes in SCs and establishes SC-GC crosstalk.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

