Endothelial progenitor cells stimulate neonatal lung angiogenesis through FOXF1-mediated activation of BMP9/ACVRL1 signaling
- PMID: 35440116
- PMCID: PMC9019054
- DOI: 10.1038/s41467-022-29746-y
Endothelial progenitor cells stimulate neonatal lung angiogenesis through FOXF1-mediated activation of BMP9/ACVRL1 signaling
Abstract
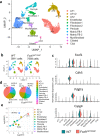
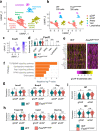
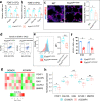
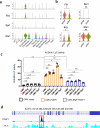
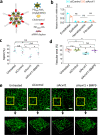
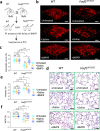
Pulmonary endothelial progenitor cells (EPCs) are critical for neonatal lung angiogenesis and represent a subset of general capillary cells (gCAPs). Molecular mechanisms through which EPCs stimulate lung angiogenesis are unknown. Herein, we used single-cell RNA sequencing to identify the BMP9/ACVRL1/SMAD1 pathway signature in pulmonary EPCs. BMP9 receptor, ACVRL1, and its downstream target genes were inhibited in EPCs from Foxf1WT/S52F mutant mice, a model of alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV). Expression of ACVRL1 and its targets were reduced in lungs of ACDMPV subjects. Inhibition of FOXF1 transcription factor reduced BMP9/ACVRL1 signaling and decreased angiogenesis in vitro. FOXF1 synergized with ETS transcription factor FLI1 to activate ACVRL1 promoter. Nanoparticle-mediated silencing of ACVRL1 in newborn mice decreased neonatal lung angiogenesis and alveolarization. Treatment with BMP9 restored lung angiogenesis and alveolarization in ACVRL1-deficient and Foxf1WT/S52F mice. Altogether, EPCs promote neonatal lung angiogenesis and alveolarization through FOXF1-mediated activation of BMP9/ACVRL1 signaling.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Generation of Pulmonary Endothelial Progenitor Cells for Cell-based Therapy Using Interspecies Mouse-Rat Chimeras.Am J Respir Crit Care Med. 2021 Aug 1;204(3):326-338. doi: 10.1164/rccm.202003-0758OC. Am J Respir Crit Care Med. 2021. PMID: 33705684 Free PMC article.
-
The S52F FOXF1 Mutation Inhibits STAT3 Signaling and Causes Alveolar Capillary Dysplasia.Am J Respir Crit Care Med. 2019 Oct 15;200(8):1045-1056. doi: 10.1164/rccm.201810-1897OC. Am J Respir Crit Care Med. 2019. PMID: 31199666 Free PMC article.
-
Nanoparticle Delivery of STAT3 Alleviates Pulmonary Hypertension in a Mouse Model of Alveolar Capillary Dysplasia.Circulation. 2021 Aug 17;144(7):539-555. doi: 10.1161/CIRCULATIONAHA.121.053980. Epub 2021 Jun 11. Circulation. 2021. PMID: 34111939 Free PMC article.
-
[Alveolar capillary dysplasia with misalignment of the pulmonary veins: a case report and literature review].Zhonghua Er Ke Za Zhi. 2020 Oct 2;58(10):838-842. doi: 10.3760/cma.j.cn112140-20200427-00441. Zhonghua Er Ke Za Zhi. 2020. PMID: 32987465 Review. Chinese.
-
Therapeutic Potential of Endothelial Progenitor Cells in Pulmonary Diseases.Am J Respir Cell Mol Biol. 2021 Nov;65(5):473-488. doi: 10.1165/rcmb.2021-0152TR. Am J Respir Cell Mol Biol. 2021. PMID: 34293272 Free PMC article. Review.
Cited by
-
Macrophage extracellular vesicle-packaged miR-23a-3p impairs maintenance and angiogenic capacity of human endothelial progenitor cells in neonatal hyperoxia-induced lung injury.Stem Cell Res Ther. 2024 Sep 11;15(1):295. doi: 10.1186/s13287-024-03920-z. Stem Cell Res Ther. 2024. PMID: 39256862 Free PMC article.
-
ALK1 Signaling in Human Cardiac Progenitor Cells Promotes a Pro-angiogenic Secretome.J Cell Signal. 2024;5(3):122-142. doi: 10.33696/signaling.5.119. J Cell Signal. 2024. PMID: 39430425 Free PMC article.
-
Single Cell Multiomics Identifies Cells and Genetic Networks Underlying Alveolar Capillary Dysplasia.Am J Respir Crit Care Med. 2023 Sep 15;208(6):709-725. doi: 10.1164/rccm.202210-2015OC. Am J Respir Crit Care Med. 2023. PMID: 37463497 Free PMC article.
-
Link between mutations in ACVRL1 and PLA2G4A genes and chronic intestinal ulcers: A case report and review of literature.World J Gastrointest Surg. 2024 Mar 27;16(3):932-943. doi: 10.4240/wjgs.v16.i3.932. World J Gastrointest Surg. 2024. PMID: 38577076 Free PMC article.
-
Endothelial to mesenchymal transition in neonatal hyperoxic lung injury: role of sex as a biological variable.Physiol Genomics. 2023 Aug 1;55(8):345-354. doi: 10.1152/physiolgenomics.00037.2023. Epub 2023 Jul 3. Physiol Genomics. 2023. PMID: 37395632 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

