Human serum triggers antibiotic tolerance in Staphylococcus aureus
- PMID: 35440121
- PMCID: PMC9018823
- DOI: 10.1038/s41467-022-29717-3
Human serum triggers antibiotic tolerance in Staphylococcus aureus
Abstract
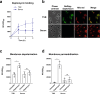
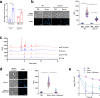
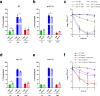
Staphylococcus aureus frequently causes infections that are challenging to treat, leading to high rates of persistent and relapsing infection. Here, to understand how the host environment influences treatment outcomes, we study the impact of human serum on staphylococcal antibiotic susceptibility. We show that serum triggers a high degree of tolerance to the lipopeptide antibiotic daptomycin and several other classes of antibiotic. Serum-induced daptomycin tolerance is due to two independent mechanisms. Firstly, the host defence peptide LL-37 induces tolerance by triggering the staphylococcal GraRS two-component system, leading to increased peptidoglycan accumulation. Secondly, GraRS-independent increases in membrane cardiolipin abundance are required for full tolerance. When both mechanisms are blocked, S. aureus incubated in serum is as susceptible to daptomycin as when grown in laboratory media. Our work demonstrates that host factors can significantly modulate antibiotic susceptibility via diverse mechanisms, and combination therapy may provide a way to mitigate this.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Public Health England. Annual epidemiological commentary: bacteraemia, MSSA bacteraemia and C. difficile infections, up to and including financial year April 2018 to March 2019. 1–88 (2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

