Multilayered regulations of alternative splicing, NMD, and protein stability control temporal induction and tissue-specific expression of TRIM46 during axon formation
- PMID: 35440129
- PMCID: PMC9019110
- DOI: 10.1038/s41467-022-29786-4
Multilayered regulations of alternative splicing, NMD, and protein stability control temporal induction and tissue-specific expression of TRIM46 during axon formation
Abstract
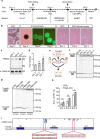
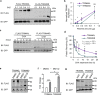
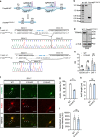
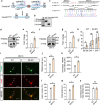
The gene regulation underlying axon formation and its exclusiveness to neurons remains elusive. TRIM46 is postulated to determine axonal fate. We show Trim46 mRNA is expressed before axonogenesis, but TRIM46 protein level is inhibited by alternative splicing of two cassette exons coupled separately to stability controls of Trim46 mRNA and proteins, effectively inducing functional knockout of TRIM46 proteins. Exon 8 inclusion causes nonsense-mediated mRNA decay of Trim46 transcripts. PTBP2-mediated exon 10 skipping produces transcripts encoding unstable TRIM46 proteins. During axonogenesis, transcriptional activation, decreased exon 8 inclusion, and enhanced exon 10 inclusion converge to increase TRIM46 proteins, leading to its neural-specific expression. Genetic deletion of these exons alters TRIM46 protein levels and shows TRIM46 is instructive though not always required for AnkG localization nor a determinant of AnkG density. Therefore, two concurrently but independently regulated alternative exons orchestrate the temporal induction and tissue-specific expression of TRIM46 proteins to mediate axon formation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Alternative splicing coupled to nonsense-mediated decay coordinates downregulation of non-neuronal genes in developing mouse neurons.Genome Biol. 2024 Jun 20;25(1):162. doi: 10.1186/s13059-024-03305-8. Genome Biol. 2024. PMID: 38902825 Free PMC article.
-
Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression.Int J Dev Neurosci. 2016 Dec;55:102-108. doi: 10.1016/j.ijdevneu.2016.03.003. Epub 2016 Mar 8. Int J Dev Neurosci. 2016. PMID: 26968265 Free PMC article. Review.
-
Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity.Nat Commun. 2017 Aug 21;8(1):306. doi: 10.1038/s41467-017-00370-5. Nat Commun. 2017. PMID: 28824175 Free PMC article.
-
Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay.Int J Mol Sci. 2020 Dec 10;21(24):9424. doi: 10.3390/ijms21249424. Int J Mol Sci. 2020. PMID: 33321981 Free PMC article. Review.
-
Axonogenesis Is Coordinated by Neuron-Specific Alternative Splicing Programming and Splicing Regulator PTBP2.Neuron. 2019 Feb 20;101(4):690-706.e10. doi: 10.1016/j.neuron.2019.01.022. Epub 2019 Feb 4. Neuron. 2019. PMID: 30733148 Free PMC article.
Cited by
-
Alternative splicing in neurodegenerative disease and the promise of RNA therapies.Nat Rev Neurosci. 2023 Aug;24(8):457-473. doi: 10.1038/s41583-023-00717-6. Epub 2023 Jun 19. Nat Rev Neurosci. 2023. PMID: 37336982 Review.
-
TRIM Proteins and Antiviral Microtubule Reorganization: A Novel Component in Innate Immune Responses?Viruses. 2024 Aug 20;16(8):1328. doi: 10.3390/v16081328. Viruses. 2024. PMID: 39205302 Free PMC article. Review.
-
Positive Selection and Duplication of Bat TRIM Family Proteins.Viruses. 2023 Mar 29;15(4):875. doi: 10.3390/v15040875. Viruses. 2023. PMID: 37112854 Free PMC article.
-
Translatome analysis in acute ischemic stroke: Astrocytes and microglia exhibit differences in poststroke alternative splicing of expressed transcripts.FASEB J. 2024 Aug 15;38(15):e23855. doi: 10.1096/fj.202400341R. FASEB J. 2024. PMID: 39096134 Free PMC article.
-
mRNA isoform balance in neuronal development and disease.Wiley Interdiscip Rev RNA. 2023 May-Jun;14(3):e1762. doi: 10.1002/wrna.1762. Epub 2022 Sep 19. Wiley Interdiscip Rev RNA. 2023. PMID: 36123820 Free PMC article. Review.

