ESR1 mutant breast cancers show elevated basal cytokeratins and immune activation
- PMID: 35440136
- PMCID: PMC9019037
- DOI: 10.1038/s41467-022-29498-9
ESR1 mutant breast cancers show elevated basal cytokeratins and immune activation
Abstract
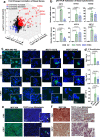
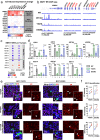
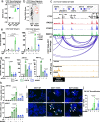
Estrogen receptor alpha (ER/ESR1) is frequently mutated in endocrine resistant ER-positive (ER+) breast cancer and linked to ligand-independent growth and metastasis. Despite the distinct clinical features of ESR1 mutations, their role in intrinsic subtype switching remains largely unknown. Here we find that ESR1 mutant cells and clinical samples show a significant enrichment of basal subtype markers, and six basal cytokeratins (BCKs) are the most enriched genes. Induction of BCKs is independent of ER binding and instead associated with chromatin reprogramming centered around a progesterone receptor-orchestrated insulated neighborhood. BCK-high ER+ primary breast tumors exhibit a number of enriched immune pathways, shared with ESR1 mutant tumors. S100A8 and S100A9 are among the most induced immune mediators and involve in tumor-stroma paracrine crosstalk inferred by single-cell RNA-seq from metastatic tumors. Collectively, these observations demonstrate that ESR1 mutant tumors gain basal features associated with increased immune activation, encouraging additional studies of immune therapeutic vulnerabilities.
© 2022. The Author(s).
Conflict of interest statement
S.Oe and A.V.L. receive research support from AstraZeneca PLC. A.V.L. is employee and consultant with UPMC Enterprises, and member of the Scientific Advisory Board, Stockholder and receives compensation from Ocean Genomics. Tsinghua University paid the stipend of University of Pittsburgh-affiliated foreign scholar Yang Wu from Tsinghua University. D.A.A.V. is cofounder and stock holder of Novasenta, Potenza, Tizona, Trishula; stock holder of Oncorus, Werewolf, Apeximmune; patents licensed and royalties of Astellas, BMS, Novasenta; scientific advisory board member—Tizona, Werewolf, F-Star, Bicara, Apeximmune; consultant of Astellas, BMS, Almirall, Incyte, G1 Therapeutics; research funding of BMS, Astellas, and Novasenta. The remaining authors declare no competing financial interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

