Distinct Survival, Growth Lag, and rRNA Degradation Kinetics during Long-Term Starvation for Carbon or Phosphate
- PMID: 35440180
- PMCID: PMC9241543
- DOI: 10.1128/msphere.01006-21
Distinct Survival, Growth Lag, and rRNA Degradation Kinetics during Long-Term Starvation for Carbon or Phosphate
Abstract
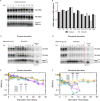
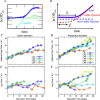
The stationary phase is the general term for the state a bacterial culture reaches when no further increase in cell mass occurs due to exhaustion of nutrients in the growth medium. Depending on the type of nutrient that is first depleted, the metabolic state of the stationary phase cells may vary greatly, and the subsistence strategies that best support cell survival may differ. As ribosomes play a central role in bacterial growth and energy expenditure, ribosome preservation is a key element of such strategies. To investigate the degree of ribosome preservation during long-term starvation, we compared the dynamics of rRNA levels of carbon-starved and phosphorus-starved Escherichia coli cultures for up to 28 days. The starved cultures' contents of full-length 16S and 23S rRNA decreased as the starvation proceeded in both cases, and phosphorus starvation resulted in much more rapid rRNA degradation than carbon starvation. Bacterial survival and regrowth kinetics were also quantified. Upon replenishment of the nutrient in question, carbon-starved cells resumed growth faster than cells starved for phosphate for the equivalent amount of time, and for both conditions, the lag time increased with the starvation time. While these results are in accordance with the hypothesis that cells with a larger ribosome pool recover more readily upon replenishment of nutrients, we also observed that the lag time kept increasing with increasing starvation time, also when the amount of rRNA per viable cell remained constant, highlighting that lag time is not a simple function of ribosome content under long-term starvation conditions. IMPORTANCE The exponential growth of bacterial populations is punctuated by long or short periods of starvation lasting from the point of nutrient exhaustion until nutrients are replenished. To understand the consequences of long-term starvation for Escherichia coli cells, we performed month-long carbon and phosphorus starvation experiments and measured three key phenotypes of the cultures, namely, the survival of the cells, the time needed for them to resume growth after nutrient replenishment, and the levels of intact rRNA preserved in the cultures. The starved cultures' concentration of rRNA dropped with starvation time, as did cell survival, while the lag time needed for regrowth increased. While all three phenotypes were more severely affected during starvation for phosphorus than for carbon, our results demonstrate that neither survival nor lag time is correlated with ribosome content in a straightforward manner.
Keywords: Escherichia coli; bacterial stress response; lag time; nutrient starvation; ribosomal RNA; stable RNA degradation; stationary phase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Maaløe O. 1979. Regulation of the protein-synthesizing machinery: ribosomes, tRNA, factors, and so on, p 487–542. In Goldberger RF (ed), Biological regulation and development. Springer, Boston, MA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

