Differential effects of macrophage subtypes on SARS-CoV-2 infection in a human pluripotent stem cell-derived model
- PMID: 35440562
- PMCID: PMC9018716
- DOI: 10.1038/s41467-022-29731-5
Differential effects of macrophage subtypes on SARS-CoV-2 infection in a human pluripotent stem cell-derived model
Abstract
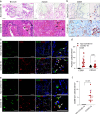
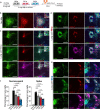
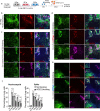
Dysfunctional immune responses contribute critically to the progression of Coronavirus Disease-2019 (COVID-19), with macrophages as one of the main cell types involved. It is urgent to understand the interactions among permissive cells, macrophages, and the SARS-CoV-2 virus, thereby offering important insights into effective therapeutic strategies. Here, we establish a lung and macrophage co-culture system derived from human pluripotent stem cells (hPSCs), modeling the host-pathogen interaction in SARS-CoV-2 infection. We find that both classically polarized macrophages (M1) and alternatively polarized macrophages (M2) have inhibitory effects on SARS-CoV-2 infection. However, M1 and non-activated (M0) macrophages, but not M2 macrophages, significantly up-regulate inflammatory factors upon viral infection. Moreover, M1 macrophages suppress the growth and enhance apoptosis of lung cells. Inhibition of viral entry using an ACE2 blocking antibody substantially enhances the activity of M2 macrophages. Our studies indicate differential immune response patterns in distinct macrophage phenotypes, which could lead to a range of COVID-19 disease severity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

