Astroblastomas exhibit radial glia stem cell lineages and differential expression of imprinted and X-inactivation escape genes
- PMID: 35440587
- PMCID: PMC9018799
- DOI: 10.1038/s41467-022-29302-8
Astroblastomas exhibit radial glia stem cell lineages and differential expression of imprinted and X-inactivation escape genes
Erratum in
-
Author Correction: Astroblastomas exhibit radial glia stem cell lineages and differential expression of imprinted and X-inactivation escape genes.Nat Commun. 2023 Jan 31;14(1):507. doi: 10.1038/s41467-023-36255-z. Nat Commun. 2023. PMID: 36720866 Free PMC article. No abstract available.
Abstract
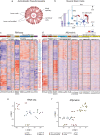
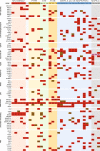
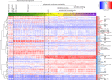
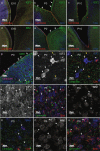
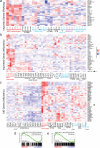
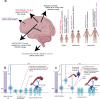
Astroblastomas (ABs) are rare brain tumors of unknown origin. We performed an integrative genetic and epigenetic analysis of AB-like tumors. Here, we show that tumors traceable to neural stem/progenitor cells (radial glia) that emerge during early to later brain development occur in children and young adults, respectively. Tumors with MN1-BEND2 fusion appear to present exclusively in females and exhibit overexpression of genes expressed prior to 25 post-conception weeks (pcw), including genes enriched in early ventricular zone radial glia and ependymal tumors. Other, histologically classic ABs overexpress or harbor mutations of mitogen-activated protein kinase pathway genes, outer and truncated radial glia genes, and genes expressed after 25 pcw, including neuronal and astrocyte markers. Findings support that AB-like tumors arise in the context of epigenetic and genetic changes in neural progenitors. Selective gene fusion, variable imprinting and/or chromosome X-inactivation escape resulting in biallelic overexpression may contribute to female predominance of AB molecular subtypes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bailey P, Bucy PC. Astroblastomas of the brain. Acta Psychiatr. Scand. 1930;5:439–461. doi: 10.1111/j.1600-0447.1930.tb08230.x. - DOI
-
- Aldape K. D., Rosenblum M. K. Astroblastoma. In: WHO Classification of Tumours of the Central Nervous System (eds Louis, D. N., Ohgaki, H., Wiestler, O. D. & Cavenee, W. K.). International Agency for Research on Cancer (2016).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

