Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF
- PMID: 35440598
- PMCID: PMC9019034
- DOI: 10.1038/s41467-022-29696-5
Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF
Abstract
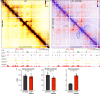
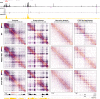
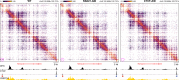
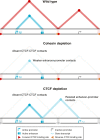
Enhancers and promoters predominantly interact within large-scale topologically associating domains (TADs), which are formed by loop extrusion mediated by cohesin and CTCF. However, it is unclear whether complex chromatin structures exist at sub-kilobase-scale and to what extent fine-scale regulatory interactions depend on loop extrusion. To address these questions, we present an MNase-based chromosome conformation capture (3C) approach, which has enabled us to generate the most detailed local interaction data to date (20 bp resolution) and precisely investigate the effects of cohesin and CTCF depletion on chromatin architecture. Our data reveal that cis-regulatory elements have distinct internal nano-scale structures, within which local insulation is dependent on CTCF, but which are independent of cohesin. In contrast, we find that depletion of cohesin causes a subtle reduction in longer-range enhancer-promoter interactions and that CTCF depletion can cause rewiring of regulatory contacts. Together, our data show that loop extrusion is not essential for enhancer-promoter interactions, but contributes to their robustness and specificity and to precise regulation of gene expression.
© 2022. The Author(s).
Conflict of interest statement
J.O.J.D. is a co-founder of Nucleome Therapeutics and provides consultancy to the company. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

