Loss of vascular endothelial notch signaling promotes spontaneous formation of tertiary lymphoid structures
- PMID: 35440634
- PMCID: PMC9018798
- DOI: 10.1038/s41467-022-29701-x
Loss of vascular endothelial notch signaling promotes spontaneous formation of tertiary lymphoid structures
Abstract
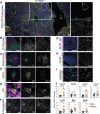
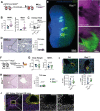
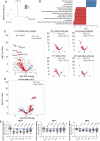
Tertiary lymphoid structures (TLS) are lymph node-like immune cell clusters that emerge during chronic inflammation in non-lymphoid organs like the kidney, but their origin remains not well understood. Here we show, using conditional deletion strategies of the canonical Notch signaling mediator Rbpj, that loss of endothelial Notch signaling in adult mice induces the spontaneous formation of bona fide TLS in the kidney, liver and lung, based on molecular, cellular and structural criteria. These TLS form in a stereotypical manner around parenchymal arteries, while secondary lymphoid structures remained largely unchanged. This effect is mediated by endothelium of blood vessels, but not lymphatics, since a lymphatic endothelial-specific targeting strategy did not result in TLS formation, and involves loss of arterial specification and concomitant acquisition of a high endothelial cell phenotype, as shown by transcriptional analysis of kidney endothelial cells. This indicates a so far unrecognized role for vascular endothelial cells and Notch signaling in TLS initiation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

