Template directed synthesis of antibody Fc conjugates with concomitant ligand release
- PMID: 35440989
- PMCID: PMC8985508
- DOI: 10.1039/d1sc06182h
Template directed synthesis of antibody Fc conjugates with concomitant ligand release
Abstract
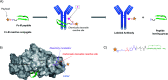
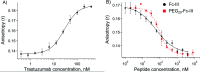
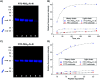
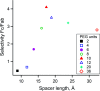
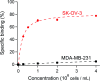
Antibodies are an attractive therapeutic modality for cancer treatment as they allow the increase of the treatment response rate and avoid the severe side effects of chemotherapy. Notwithstanding the strong benefit of antibodies, the efficacy of anti-cancer antibodies can dramatically vary among patients and ultimately result in no response to the treatment. Here, we have developed a novel means to regioselectively label the Fc domain of any therapeutic antibody with a radionuclide chelator in a single step chemistry, with the aim to study by SPECT/CT imaging if the radiolabeled antibody is capable of targeting cancer cells in vivo. A Fc-III peptide was used as bait to bring a carbonate electrophilic site linked to a metal chelator and to a carboxyphenyl leaving group in close proximity with an antibody Fc nucleophile amino acid (K317), thereby triggering the covalent linkage of the chelator to the antibody lysine, with the concomitant release of the carboxyphenyl Fc-III ligand. Using CHX-A''-DTPA, we radiolabeled trastuzumab with indium-111 and showed in biodistribution and imaging experiments that the antibody accumulated successfully in the SK-OV-3 xenograft tumour implanted in mice. We found that our methodology leads to homogeneous conjugation of CHX-A''-DTPA to the antibody, and confirmed that the Fc domain can be selectively labeled at K317, with a minor level of unspecific labeling on the Fab domain. The present method can be developed as a clinical diagnostic tool to predict the success of the therapy. Furthermore, our Fc-III one step chemistry concept paves the way to a broad array of other applications in antibody bioengineering.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare. V. P, L. M, F. L, P. G, J.-M. S, and O. N are coinventors of application WO2021110860 A1 filed on 3 December 2019, entitled “Reactive conjugates”.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

