pH-EVD: A pH-Paper-Based Extraction and Visual Detection System for Instrument-Free SARS-CoV-2 Diagnostics
- PMID: 35441159
- PMCID: PMC9011642
- DOI: 10.1002/anbr.202100101
pH-EVD: A pH-Paper-Based Extraction and Visual Detection System for Instrument-Free SARS-CoV-2 Diagnostics
Abstract
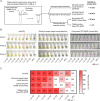
The ongoing pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused millions of deaths worldwide. However, most SARS-CoV-2 detection methods depend on time-consuming sample preparation and large detection instruments. Herein, a method employing nonbleeding pH paper to achieve both RNA extraction and visual isothermal amplification is proposed, enabling rapid, instrument-free SARS-CoV-2 detection. By taking advantage of capillary forces, pH-paper-based RNA extraction can be accomplished within 1 min without need for any equipment. Further, the pH paper can mediate dye-free visual isothermal amplification detection. In less than a 46-min sample-to-answer time, pH-paper-based extraction and visual detection (termed pH-EVD) can consistently detect 1200 genome equivalents per microliter of SARS-CoV-2 in saliva, which is comparable to TaqMan probe-based quantitative reverse transcription PCR (RT-qPCR). Through coupling with a chemically heated incubator called a smart cup, the instrument-free, pH-EVD-based SARS-CoV-2 detection method on 30 nasopharyngeal swab samples and 33 contrived saliva samples is clinically validated. Thus, the pH-EVD method provides simple, rapid, reliable, low-cost, and instrument-free SARS-CoV-2 detection and has the potential to streamline onsite COVID-19 diagnostics.
Keywords: RNA extraction; SARS-CoV-2; instrument-free onsite diagnostics; nonbleeding pH paper; visual isothermal amplification detection.
© 2021 The Authors. Advanced NanoBiomed Research published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Ultrasensitive Detection of SARS-CoV-2 in Saliva and Viral Transport Medium Clinical Samples.Anal Chem. 2021 Jun 8;93(22):7797-7807. doi: 10.1021/acs.analchem.0c05170. Epub 2021 May 25. Anal Chem. 2021. PMID: 34033472
-
CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology.J Appl Microbiol. 2022 Aug;133(2):410-421. doi: 10.1111/jam.15571. Epub 2022 Apr 18. J Appl Microbiol. 2022. PMID: 35396760 Free PMC article.
-
Development and Clinical Application of a Rapid and Sensitive Loop-Mediated Isothermal Amplification Test for SARS-CoV-2 Infection.mSphere. 2020 Aug 26;5(4):e00808-20. doi: 10.1128/mSphere.00808-20. mSphere. 2020. PMID: 32848011 Free PMC article.
-
Isothermal amplification-based assays for rapid and sensitive detection of severe acute respiratory syndrome coronavirus 2: Opportunities and recent developments.Rev Med Virol. 2022 Mar;32(2):e2274. doi: 10.1002/rmv.2274. Epub 2021 Jul 3. Rev Med Virol. 2022. PMID: 34216498 Free PMC article. Review.
-
Different Respiratory Samples for COVID-19 Detection by Standard and Direct Quantitative RT-PCR: A Literature Review.Iran J Pharm Res. 2021 Summer;20(3):285-299. doi: 10.22037/ijpr.2021.115458.15383. Iran J Pharm Res. 2021. PMID: 34903989 Free PMC article. Review.
Cited by
-
Paper-Based Biosensors for the Detection of Nucleic Acids from Pathogens.Biosensors (Basel). 2022 Nov 29;12(12):1094. doi: 10.3390/bios12121094. Biosensors (Basel). 2022. PMID: 36551061 Free PMC article. Review.
-
Sample preparation and detection methods in point-of-care devices towards future at-home testing.Lab Chip. 2024 Jul 23;24(15):3626-3650. doi: 10.1039/d3lc00943b. Lab Chip. 2024. PMID: 38952234 Free PMC article. Review.
-
Biosensors based on potent miniprotein binder for sensitive testing of SARS-CoV‑2 variants of concern.Mikrochim Acta. 2023 Dec 18;191(1):38. doi: 10.1007/s00604-023-06113-2. Mikrochim Acta. 2023. PMID: 38110824
-
MiR-19-loaded oxidative stress-relief microgels with immunomodulatory and regeneration functions to reduce cardiac remodeling after myocardial infarction.Bioact Mater. 2025 Feb 13;48:43-54. doi: 10.1016/j.bioactmat.2025.02.004. eCollection 2025 Jun. Bioact Mater. 2025. PMID: 40303965 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
