Plant biosynthetic gene clusters in the context of metabolic evolution
- PMID: 35441651
- PMCID: PMC9298681
- DOI: 10.1039/d2np00005a
Plant biosynthetic gene clusters in the context of metabolic evolution
Abstract
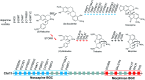
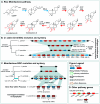
Covering: up to 2022Plants produce a wide range of structurally and biosynthetically diverse natural products to interact with their environment. These specialised metabolites typically evolve in limited taxonomic groups presumably in response to specific selective pressures. With the increasing availability of sequencing data, it has become apparent that in many cases the genes encoding biosynthetic enzymes for specialised metabolic pathways are not randomly distributed on the genome. Instead they are physically linked in structures such as arrays, pairs and clusters. The exact function of these clusters is debated. In this review we take a broad view of gene arrangement in plant specialised metabolism, examining types of structures and variation. We discuss the evolution of biosynthetic gene clusters in the wider context of metabolism, populations and epigenetics. Finally, we synthesise our observations to propose a new hypothesis for biosynthetic gene cluster formation in plants.
Conflict of interest statement
There are no conflicts to declare.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

