A loss-of-function IFNAR1 allele in Polynesia underlies severe viral diseases in homozygotes
- PMID: 35442418
- PMCID: PMC9026234
- DOI: 10.1084/jem.20220028
A loss-of-function IFNAR1 allele in Polynesia underlies severe viral diseases in homozygotes
Abstract
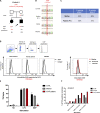
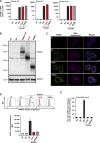
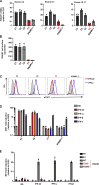
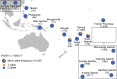
Globally, autosomal recessive IFNAR1 deficiency is a rare inborn error of immunity underlying susceptibility to live attenuated vaccine and wild-type viruses. We report seven children from five unrelated kindreds of western Polynesian ancestry who suffered from severe viral diseases. All the patients are homozygous for the same nonsense IFNAR1 variant (p.Glu386*). This allele encodes a truncated protein that is absent from the cell surface and is loss-of-function. The fibroblasts of the patients do not respond to type I IFNs (IFN-α2, IFN-ω, or IFN-β). Remarkably, this IFNAR1 variant has a minor allele frequency >1% in Samoa and is also observed in the Cook, Society, Marquesas, and Austral islands, as well as Fiji, whereas it is extremely rare or absent in the other populations tested, including those of the Pacific region. Inherited IFNAR1 deficiency should be considered in individuals of Polynesian ancestry with severe viral illnesses.
© 2022 Bastard et al.
Conflict of interest statement
Disclosures: A.J. Mentzer reports grants from the National Institutes for Health Research during the conduct of the study. No other disclosures were reported.
Figures




References
-
- Abolhassani, H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., Aranda-Guillen M., Sardh F., Zuo F., Zhang P., et al. 2022. Inherited IFNAR1 deficiency in a child with both critical COVID-19 pneumonia and multisystem inflammatory syndrome. J. Clin. Immunol. 1–13. 10.1007/s10875-022-01215-7 - DOI - PMC - PubMed
-
- Addison, D.J., and Matisoo-Smith E.. 2010. Rethinking Polynesians origins: A West-Polynesia triple-I model. Archaeol Ocean. 45:1–12. 10.1002/j.1834-4453.2010.tb00072.x - DOI
-
- Amatuni, G.S., Currier R.J., Church J.A., Bishop T., Grimbacher E., Nguyen A.A.-C., Agarwal-Hashmi R., Aznar C.P., Butte M.J., Cowan M.J., et al. 2019. Newborn screening for severe combined immunodeficiency and T-cell lymphopenia in California, 2010-2017. Pediatrics. 143:e20182300. 10.1542/peds.2018-2300 - DOI - PMC - PubMed
-
- Bastard, P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.-H., Eto S., Garcia-Prat M., et al. 2021a. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci. Immunol. 6:eabl4340. 10.1126/sciimmunol.abl4340 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- HHSN268201100037C/HL/NHLBI NIH HHS/United States
- R01AI088364/NH/NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- S10 OD018521/OD/NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- HHSN268201500016C/HL/NHLBI NIH HHS/United States
- R01 AI088364/AI/NIAID NIH HHS/United States
- R01 HL093093/HL/NHLBI NIH HHS/United States
- MR/N028937/1/MRC_/Medical Research Council/United Kingdom
- U24 HG008956/HG/NHGRI NIH HHS/United States
- UM1 HG006504/HG/NHGRI NIH HHS/United States
- R01 HL133040/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- UM1HG006504/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UL1 RR024143/RR/NCRR NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- R01 AI163029/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

