Multiscale affinity maturation simulations to elicit broadly neutralizing antibodies against HIV
- PMID: 35442968
- PMCID: PMC9020693
- DOI: 10.1371/journal.pcbi.1009391
Multiscale affinity maturation simulations to elicit broadly neutralizing antibodies against HIV
Abstract
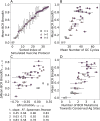
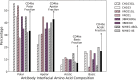
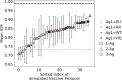
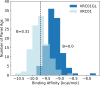
The design of vaccines against highly mutable pathogens, such as HIV and influenza, requires a detailed understanding of how the adaptive immune system responds to encountering multiple variant antigens (Ags). Here, we describe a multiscale model of B cell receptor (BCR) affinity maturation that employs actual BCR nucleotide sequences and treats BCR/Ag interactions in atomistic detail. We apply the model to simulate the maturation of a broadly neutralizing Ab (bnAb) against HIV. Starting from a germline precursor sequence of the VRC01 anti-HIV Ab, we simulate BCR evolution in response to different vaccination protocols and different Ags, which were previously designed by us. The simulation results provide qualitative guidelines for future vaccine design and reveal unique insights into bnAb evolution against the CD4 binding site of HIV. Our model makes possible direct comparisons of simulated BCR populations with results of deep sequencing data, which will be explored in future applications.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: AKC is a consultant (titled “academic partner”) with Flagship Pioneering and its affiliated companies, Apriori Bio and FL72, and serves on the Strategic Oversight Boards of the latter two companies.
Figures

References
-
- UNAIDS Data 2018 | HIV/AIDS Data Hub for the Asia-Pacific Region. [Cited 2021 Aug 4]. Available from: https://www.aidsdatahub.org/resource/unaids-data-2018.
-
- Influenza (Seasonal). [Cited 2021 Aug 4]. Available from: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

