Inhibitory co-transmission from midbrain dopamine neurons relies on presynaptic GABA uptake
- PMID: 35443174
- PMCID: PMC9097974
- DOI: 10.1016/j.celrep.2022.110716
Inhibitory co-transmission from midbrain dopamine neurons relies on presynaptic GABA uptake
Abstract
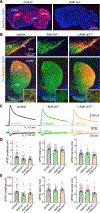
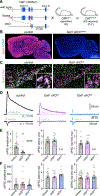
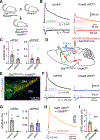
Dopamine (DA)-releasing neurons in the substantia nigra pars compacta (SNcDA) inhibit target cells in the striatum through postsynaptic activation of γ-aminobutyric acid (GABA) receptors. However, the molecular mechanisms responsible for GABAergic signaling remain unclear, as SNcDA neurons lack enzymes typically required to produce GABA or package it into synaptic vesicles. Here, we show that aldehyde dehydrogenase 1a1 (Aldh1a1), an enzyme proposed to function as a GABA synthetic enzyme in SNcDA neurons, does not produce GABA for synaptic transmission. Instead, we demonstrate that SNcDA axons obtain GABA exclusively through presynaptic uptake using the membrane GABA transporter Gat1 (encoded by Slc6a1). GABA is then packaged for vesicular release using the vesicular monoamine transporter Vmat2. Our data therefore show that presynaptic transmitter recycling can substitute for de novo GABA synthesis and that Vmat2 contributes to vesicular GABA transport, expanding the range of molecular mechanisms available to neurons to support inhibitory synaptic communication.
Keywords: Aldh1a1; CP; GABA; Gat1; Neuroscience; SLC18A2; Slc6a1; VMAT2; basal ganglia; dopamine; striatum; synaptic transmission.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
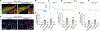
References
-
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding R, Ji FY, Kanbara N, Kuzume H, Sanbo M, Yagi T, et al. (1996). Mice lacking the 65 kDa isoform of glutamic acid decarboxylase (GAD65) maintain normal levels of GAD67 and GABA in their brains but are susceptible to seizures. Biochem. Biophys. Res. Commun. 229, 891–895. - PubMed
-
- Bäckman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, and Tomac AC (2006). Characterization of a mouse strain expressing Cre recombinase from the 3’ untranslated region of the dopamine transporter locus. Genesis 44, 383–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

