Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle
- PMID: 35444262
- PMCID: PMC9296628
- DOI: 10.1038/s41396-022-01226-7
Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle
Abstract
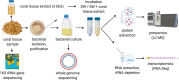
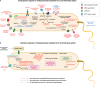
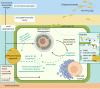
Endozoicomonas are prevalent, abundant bacterial associates of marine animals, including corals. Their role in holobiont health and functioning, however, remains poorly understood. To identify potential interactions within the coral holobiont, we characterized the novel isolate Endozoicomonas marisrubri sp. nov. 6c and assessed its transcriptomic and proteomic response to tissue extracts of its native host, the Red Sea coral Acropora humilis. We show that coral tissue extracts stimulated differential expression of genes putatively involved in symbiosis establishment via the modulation of the host immune response by E. marisrubri 6c, such as genes for flagellar assembly, ankyrins, ephrins, and serpins. Proteome analyses revealed that E. marisrubri 6c upregulated vitamin B1 and B6 biosynthesis and glycolytic processes in response to holobiont cues. Our results suggest that the priming of Endozoicomonas for a symbiotic lifestyle involves the modulation of host immunity and the exchange of essential metabolites with other holobiont members. Consequently, Endozoicomonas may play an important role in holobiont nutrient cycling and may therefore contribute to coral health, acclimatization, and adaptation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Feeley KJ, Rehm EM, Machovina B. perspective: The responses of tropical forest species to global climate change: acclimate, adapt, migrate, or go extinct? Front Biogeogr. 2012;4:69–84. doi: 10.21425/F54212621. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

