Genetic and Functional Analyses of Archaeal ATP-Dependent RNA Ligase in C/D Box sRNA Circularization and Ribosomal RNA Processing
- PMID: 35445080
- PMCID: PMC9014305
- DOI: 10.3389/fmolb.2022.811548
Genetic and Functional Analyses of Archaeal ATP-Dependent RNA Ligase in C/D Box sRNA Circularization and Ribosomal RNA Processing
Abstract
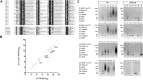
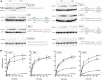
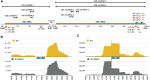
RNA ligases play important roles in repairing and circularizing RNAs post-transcriptionally. In this study, we generated an allelic knockout of ATP-dependent RNA ligase (Rnl) in the hyperthermophilic archaeon Thermococcus kodakarensis to identify its biological targets. A comparative analysis of circular RNA reveals that the Rnl-knockout strain represses circularization of C/D box sRNAs without affecting the circularization of tRNA and rRNA processing intermediates. Recombinant archaeal Rnl could circularize C/D box sRNAs with a mutation in the conserved C/D box sequence element but not when the terminal stem structures were disrupted, suggesting that proximity of the two ends could be critical for intramolecular ligation. Furthermore, T. kodakarensis accumulates aberrant RNA fragments derived from ribosomal RNA in the absence of Rnl. These results suggest that Rnl is responsible for C/D box sRNA circularization and may also play a role in ribosomal RNA processing.
Keywords: C/D box sRNAs; RNA ligase; circular RNA; rRNA processing; thermococcus kodakarensis KOD1.
Copyright © 2022 Liu, Takagi, Sugijanto, Nguyen, Hirata, Hori and Ho.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

