The genomic landscape of pediatric renal cell carcinomas
- PMID: 35445187
- PMCID: PMC9014386
- DOI: 10.1016/j.isci.2022.104167
The genomic landscape of pediatric renal cell carcinomas
Abstract
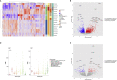
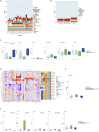
Pediatric renal cell carcinomas (RCC) differ from their adult counterparts not only in histologic subtypes but also in clinical characteristics and outcome. However, the underlying biology is still largely unclear. For this reason, we performed whole-exome and transcriptome sequencing analyses on a cohort of 25 pediatric RCC patients with various histologic subtypes, including 10 MiT family translocation (MiT) and 10 papillary RCCs. In this cohort of pediatric RCC, we find only limited genomic overlap with adult RCC, even within the same histologic subtype. Recurrent somatic mutations in genes not previously reported in RCC were detected, such as in CCDC168, PLEKHA1, VWF, and MAP3K9. Our papillary pediatric RCCs, which represent the largest cohort to date with comprehensive molecular profiling in this age group, appeared as a distinct genomic subtype differing in terms of gene mutations and gene expression patterns not only from MiT-RCC but also from their adult counterparts.
Keywords: Cancer; Genomics.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Albiges L., Guegan J., Le Formal A., Verkarre V., Rioux-Leclercq N., Sibony M., Bernhard J.C., Camparo P., Merabet Z., Molinie V., et al. MET is a potential target across all papillary renal cell carcinomas: result from a large molecular study of pRCC with CGH array and matching gene expression array. Clin. Cancer Res. 2014;20:3411–3421. - PubMed
-
- Andrews L.P., Yano H., Vignali D.A.A. Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat. Immunol. 2019;20:1425–1434. - PubMed
-
- Ayala-Sarmiento A.E., Martinez-Fong D., Segovia J. The internalization of neurotensin by the low-affinity neurotensin receptors (NTSR2 and vNTSR2) activates ERK 1/2 in glioma cells and allows neurotensin-polyplex transfection of tGAS1. Cell Mol. Neurobiol. 2015;35:785–795. doi: 10.1007/s10571-015-0172-z. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

