Molecular Epidemiological Characteristics of Mycobacterium abscessus Complex Derived from Non-Cystic Fibrosis Patients in Japan and Taiwan
- PMID: 35446117
- PMCID: PMC9248903
- DOI: 10.1128/spectrum.00571-22
Molecular Epidemiological Characteristics of Mycobacterium abscessus Complex Derived from Non-Cystic Fibrosis Patients in Japan and Taiwan
Abstract
Mycobacterium abscessus complex (MABC) is a group of emerging, highly antimicrobial-resistant non-tuberculous mycobacteria. Specific MABC clones are spreading globally in patients with cystic fibrosis (CF); however, associated genomic epidemiology is lacking in East Asia, with very few patients with CF. Here, we investigated MABC populations derived from non-CF patients in Japan and Taiwan. Analysis of whole-genome sequencing data of 220 MABC isolates revealed that 112, 105, and 3 were M. abscessus subsp. abscessus (ABS), M. abscessus subsp. massiliense (MAS), and M. abscessus subsp. bolletii (BOL), respectively. Moreover, >50% of ABS and >70% of MAS were related to four predominant clones in the region. Known mutations conferring macrolide resistance were rare (1.4%) and were not enriched in the predominant clones. Conversely, the macrolide-susceptible erm(41) T28C mutation was significantly enriched in one predominant ABS clone. The most predominant ABS clone was genetically related to the previously described dominant circulating clone (DCC)1 in patients with CF, whereas no isolates were related to DCC2; isolates related to DCC3 were not necessarily predominant in our sample set. We found that the erm(41) T28C mutants spread globally, and some of them reacquired the functional erm(41) gene through both point mutation and recombination. This study revealed predominant MABC clones in Japan and Taiwan and their relationship with the globally superadding clones in the patient community with CF. Our study provides insights into the genetic characteristics of globally dominant and area-specific strains isolated from patients with or without CF and differences between globally spread and regionally specific strains. IMPORTANCE Members of Mycobacterium abscessus complex (MABC) are frequently isolated from patients. Studies have reported that predominant clones of MABC (known as dominant circulating clones; DCCs) are distributed worldwide and transmitted from humans to humans in patients with cystic fibrosis (CF). However, associated genomic epidemiology has not yet been conducted in East Asia, including Japan and Taiwan, where there are only a few patients with CF. Using whole-genome sequencing data derived from non-CF patients in Japan and Taiwan, we revealed prevalent clones and the incidence of macrolide resistance-associated mutations in the MABC population in this region. We also clarified the associations between these predominant clones and DCCs in the global CF patient community. Our results would assist further studies in elucidating the genetic characteristics of strains isolated from patients with or without CF, the differences between globally spread and regionally specific strains, and the adaptive evolution of MABC within the host.
Keywords: Mycobacterium abscessus; molecular epidemiology; non-cystic fibrosis; non-tuberculous mycobacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
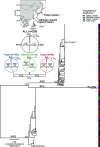


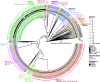



References
-
- Roux A-L, Catherinot E, Ripoll F, Soismier N, Macheras E, Ravilly S, Bellis G, Vibet M-A, Le Roux E, Lemonnier L, Gutierrez C, Vincent V, Fauroux B, Rottman M, Guillemot D, Gaillard J-L, Jean-Louis Herrmann for the OMA Group . 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J Clin Microbiol 47:4124–4128. doi: 10.1128/JCM.01257-09. - DOI - PMC - PubMed
-
- Olivier KN, Weber DJ, Wallace RJ, Faiz AR, Lee J-H, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, Edwards LJ, Chakraborti S, Knowles MR. 2003. Nontuberculous mycobacteria: I: Multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 167:828–834. doi: 10.1164/rccm.200207-678OC. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

